Impact of bacterial sRNAs in stress responses
- PMID: 29101308
- PMCID: PMC5730939
- DOI: 10.1042/BST20160363
Impact of bacterial sRNAs in stress responses
Abstract
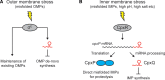
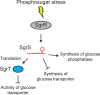
Bacterial life is harsh and involves numerous environmental and internal challenges that are perceived as stresses. Consequently, adequate responses to survive, cope with, and counteract stress conditions have evolved. In the last few decades, a class of small, non-coding RNAs (sRNAs) has been shown to be involved as key players in stress responses. This review will discuss - primarily from an enterobacterial perspective - selected stress response pathways that involve antisense-type sRNAs. These include themes of how bacteria deal with severe envelope stress, threats of DNA damage, problems with poisoning due to toxic sugar intermediates, issues of iron homeostasis, and nutrient limitation/starvation. The examples discussed highlight how stress relief can be achieved, and how sRNAs act mechanistically in regulatory circuits. For some cases, we will propose scenarios that may suggest why contributions from post-transcriptional control by sRNAs, rather than transcriptional control alone, appear to be a beneficial and universally selected feature.
Keywords: antisense mechanism; bacterial stress response; envelope stress; post-transcriptional regulation; sRNA; starvation.
© 2017 The Author(s).
Conflict of interest statement
The Authors declare that there are no competing interests associated with the manuscript.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

