In vitro selections of mammaglobin A and mammaglobin B aptamers for the recognition of circulating breast tumor cells
- PMID: 29101327
- PMCID: PMC5670216
- DOI: 10.1038/s41598-017-13751-z
In vitro selections of mammaglobin A and mammaglobin B aptamers for the recognition of circulating breast tumor cells
Abstract
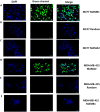
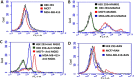
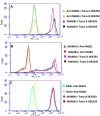
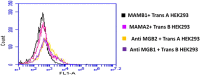
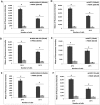
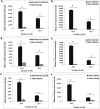
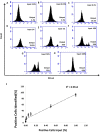
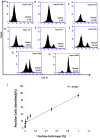
Mammaglobin B (MGB2) and mammaglobin A (MGB1) are proteins expressed in metastatic breast cancers. The early detection of circulating tumor cells (CTCs) in breast cancer patients is crucial to decrease mortality rate. Herein, novel aptamers were successfully selected and characterized against MGB2 and MGB1 proteins using a hybrid SELEX approach. The potential use of the selected aptamers in breast CTC detection was studied using spiked breast cancer cells in whole blood lysate. The results obtained from this study showed that the selected aptamers (MAMB1 and MAMA2) bind to their target breast cancer cell lines with high affinity (low nanomolar Kd values) and specificity. They also bind to their free recombinant target proteins and show minimal non-specific binding to normal and other cancer cell lines. Additionally, they were able to distinguish a low number of breast cancer cells spiked in whole blood lysate containing normal blood cells. The results obtained in this study indicate the great potential for the use of aptamers to detect MGB1 and MGB2 protein biomarkers, expressed on the surface of breast CTCs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
[Mammaglobin in peripheral blood and tumor in breast cancer patients].Biomed Khim. 2016 May;62(4):453-7. doi: 10.18097/PBMC20166204453. Biomed Khim. 2016. PMID: 27563000 Russian.
-
Detection of cytokeratin 19, human mammaglobin, and carcinoembryonic antigen-positive circulating tumor cells by three-marker reverse transcription-PCR assay and its relation to clinical outcome in early breast cancer.Int J Biol Markers. 2010 Apr-Jun;25(2):59-68. doi: 10.1177/172460081002500201. Int J Biol Markers. 2010. PMID: 20586026
-
Detection and quantification of mammaglobin in the blood of breast cancer patients: can it be useful as a potential clinical marker? Preliminary results of a GOIM (Gruppo Oncologico dell'Italia Meridionale) prospective study.Ann Oncol. 2006 Jun;17 Suppl 7:vii41-5. doi: 10.1093/annonc/mdl948. Ann Oncol. 2006. PMID: 16760290
-
Mammaglobin A: review and clinical utility.Adv Clin Chem. 2014;64:241-68. Adv Clin Chem. 2014. PMID: 24938021 Review.
-
Human mammaglobin in breast cancer: a brief review of its clinical utility.Indian J Med Res. 2014 May;139(5):675-85. Indian J Med Res. 2014. PMID: 25027076 Free PMC article. Review.
Cited by
-
Detection of Minimal Residual Disease in the Peripheral Blood of Breast Cancer Patients, with a Multi Marker (MGB-1, MGB-2, CK-19 and NY-BR-1) Assay.Breast Cancer (Dove Med Press). 2021 Nov 16;13:617-624. doi: 10.2147/BCTT.S337075. eCollection 2021. Breast Cancer (Dove Med Press). 2021. PMID: 34815711 Free PMC article.
-
Mammaglobin B may be a prognostic biomarker of uterine corpus endometrial cancer.Oncol Lett. 2020 Nov;20(5):255. doi: 10.3892/ol.2020.12118. Epub 2020 Sep 18. Oncol Lett. 2020. PMID: 32994818 Free PMC article.
-
Prognostic role of plasma mammaglobin A expression in breast carcinoma patients: a meta-analysis.Onco Targets Ther. 2018 May 30;11:3245-3255. doi: 10.2147/OTT.S156556. eCollection 2018. Onco Targets Ther. 2018. PMID: 29881297 Free PMC article. Review.
-
Cardamonin suppressed the migration, invasion, epithelial mesenchymal transition (EMT) and lung metastasis of colorectal cancer cells by down-regulating ADRB2 expression.Pharm Biol. 2022 Dec;60(1):1011-1021. doi: 10.1080/13880209.2022.2069823. Pharm Biol. 2022. PMID: 35645356 Free PMC article.
-
DNA aptamers against bacterial cells can be efficiently selected by a SELEX process using state-of-the art qPCR and ultra-deep sequencing.Sci Rep. 2020 Dec 1;10(1):20917. doi: 10.1038/s41598-020-77221-9. Sci Rep. 2020. PMID: 33262379 Free PMC article.
References
-
- Ferlay, J. et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. International Agency for Research on Cancer. Available on: http://globocan.iarc.fr (2013).
-
- U.S. Breast cancer statistics. http://www.breastcancer.org/ (2016). - PubMed
-
- Vanio, H., Bianchini, F. IARC Handbooks of Cancer Prevention, Volume 7: Breast Cancer Screening. 4–16 (IARC Press, France, 2002).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

