Autoinhibitory sterol sulfates mediate programmed cell death in a bloom-forming marine diatom
- PMID: 29101388
- PMCID: PMC5670183
- DOI: 10.1038/s41467-017-01300-1
Autoinhibitory sterol sulfates mediate programmed cell death in a bloom-forming marine diatom
Abstract
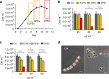
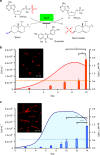
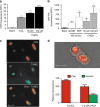
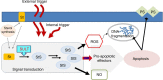
Cell mortality is a key mechanism that shapes phytoplankton blooms and species dynamics in aquatic environments. Here we show that sterol sulfates (StS) are regulatory molecules of a cell death program in Skeletonema marinoi, a marine diatom-blooming species in temperate coastal waters. The molecules trigger an oxidative burst and production of nitric oxide in a dose-dependent manner. The intracellular level of StS increases with cell ageing and ultimately leads to a mechanism of apoptosis-like death. Disrupting StS biosynthesis by inhibition of the sulfonation step significantly delays the onset of this fatal process and maintains steady growth in algal cells for several days. The autoinhibitory activity of StS demonstrates the functional significance of small metabolites in diatoms. The StS pathway provides another view on cell regulation during bloom dynamics in marine habitats and opens new opportunities for the biochemical control of mass-cultivation of microalgae.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

