Intrinsic MYH7 expression regulation contributes to tissue level allelic imbalance in hypertrophic cardiomyopathy
- PMID: 29101517
- PMCID: PMC5742120
- DOI: 10.1007/s10974-017-9486-4
Intrinsic MYH7 expression regulation contributes to tissue level allelic imbalance in hypertrophic cardiomyopathy
Abstract
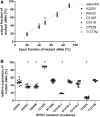
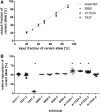
HCM, the most common inherited cardiac disease, is mainly caused by mutations in sarcomeric genes. More than a third of the patients are heterozygous for mutations in the MYH7 gene encoding for the β-myosin heavy chain. In HCM-patients, expression of the mutant and the wildtype allele can be unequal, thus leading to fractions of mutant and wildtype mRNA and protein which deviate from 1:1. This so-called allelic imbalance was detected in whole tissue samples but also in individual cells. There is evidence that the severity of HCM not only depends on the functional effect of the mutation itself, but also on the fraction of mutant protein in the myocardial tissue. Allelic imbalance has been shown to occur in a broad range of genes. Therefore, we aimed to examine whether the MYH7-alleles are intrinsically expressed imbalanced or whether the allelic imbalance is solely associated with the disease. We compared the expression of MYH7-alleles in non-HCM donors and in HCM-patients with different MYH7-missense mutations. In the HCM-patients, we identified imbalanced as well as equal expression of both alleles. Also at the protein level, allelic imbalance was determined. Most interestingly, we also discovered allelic imbalance and balance in non-HCM donors. Our findings therefore strongly indicate that apart from mutation-specific mechanisms, also non-HCM associated allelic-mRNA expression regulation may account for the allelic imbalance of the MYH7 gene in HCM-patients. Since the relative amount of mutant mRNA and protein or the extent of allelic imbalance has been associated with the severity of HCM, individual analysis of the MYH7-allelic expression may provide valuable information for the prognosis of each patient.
Keywords: Allelic imbalance; Beta-myosin; Hypertrophic cardiomyopathy; MYH7.
Figures
References
-
- Anan R, Greve G, Thierfelder L, Watkins H, McKenna WJ, Solomon S, Vecchio C, Shono H, Nakao S, Tanaka H, et al. Prognostic implications of novel beta cardiac myosin heavy chain gene mutations that cause familial hypertrophic cardiomyopathy. J Clin Invest. 1994;1:280–285. doi: 10.1172/JCI116957. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

