Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China
- PMID: 29102111
- PMCID: PMC7117260
- DOI: 10.1016/j.vetmic.2017.09.020
Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China
Abstract
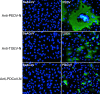
Outbreaks of diarrhea in newborn piglets without detection of transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV) and porcine deltacoronavirus (PDCoV), have been recorded in a pig farm in southern China since February 2017. Isolation and propagation of the pathogen in cell culture resulted in discovery of a novel swine enteric alphacoronavirus (tentatively named SeACoV) related to the bat coronavirus HKU2 identified in the same region a decade ago. Specific fluorescence signal was detected in Vero cells infected with SeACoV by using a positive sow serum collected in the same farm, but not by using TGEV-, PEDV- or PDCoV-specific antibody. Electron microscopy observation demonstrated that the virus particle with surface projections was 100-120nm in diameter. Complete genomic sequencing and analyses of SeACoV indicated that the extreme amino-terminal domain of the SeACoV spike (S) glycoprotein structurally similar to the domain 0 of the alphacoronavirus NL63, whereas the rest part of S structurally resembles domains B to D of the betacoronavirus. The SeACoV-S domain 0 associated with enteric tropism had an extremely high variability, harboring 75-amino-acid (aa) substitutions and a 2-aa insertion, compared to that of HKU2, which is likely responsible for the extended host range or cross-species transmission. The isolated virus was infectious in pigs when inoculated orally into 3-day-old newborn piglets, leading to clinical signs of diarrhea and fecal virus shedding. These results confirmed that it is a novel swine enteric coronavirus representing the fifth porcine coronavirus.
Keywords: Bat; Cross-species transmission; Spike glycoprotein; Swine enteric alphacoronavirus (SeACoV).
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures




References
-
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. Coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; London: 2011. pp. 806–828.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

