Is the Enzyme ACMSD a Novel Therapeutic Target in Parkinson's Disease?
- PMID: 29103054
- PMCID: PMC5676848
- DOI: 10.3233/JPD-171240
Is the Enzyme ACMSD a Novel Therapeutic Target in Parkinson's Disease?
Abstract
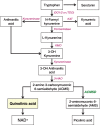
Several large genome wide association studies have identified a locus in close proximity to the gene encoding the enzyme aminocarboxymuconate-semialdehyde-decarboxylase (ACMSD) to be associated with the risk for Parkinson's disease (PD), tentatively suggesting that this enzyme might influence PD pathogenesis. Further support for this comes from the recent identification of a disease-segregating stop codon mutation in ACMSD in a family with Parkinsonism, and a missense mutation in the ACMSD gene predicted to disrupt enzyme function in an individual with typical PD. ACMSD is part of the kynurenine pathway, responsible for the catalytic breakdown of tryptophan into NAD+, generating several neuroactive metabolites in the process. The enzyme is located at a key branch-point of the pathway, limiting the production of the neurotoxin quinolinic acid, which has excitotoxic and inflammatory properties. In this review, we discuss the genetic findings in light of the functions of ACMSD and its potential involvement in PD pathogenesis.
Keywords: ACMSD; Parkinson’s disease; excitotoxicity; kynurenine pathway; neuroinflammation; oxidative stress.
Figures



Similar articles
-
The ACMSD gene, involved in tryptophan metabolism, is mutated in a family with cortical myoclonus, epilepsy, and parkinsonism.J Mol Med (Berl). 2013 Dec;91(12):1399-406. doi: 10.1007/s00109-013-1075-4. Epub 2013 Aug 20. J Mol Med (Berl). 2013. PMID: 23955123 Free PMC article.
-
Tissue expression and biochemical characterization of human 2-amino 3-carboxymuconate 6-semialdehyde decarboxylase, a key enzyme in tryptophan catabolism.FEBS J. 2007 Feb;274(3):827-40. doi: 10.1111/j.1742-4658.2007.05635.x. FEBS J. 2007. PMID: 17288562
-
Identification and expression of alpha cDNA encoding human 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase (ACMSD): a key enzyme for the tryptophan-niacine pathway and quinolinate hypothesis.Adv Exp Med Biol. 2003;527:443-53. doi: 10.1007/978-1-4615-0135-0_52. Adv Exp Med Biol. 2003. PMID: 15206762
-
Can kynurenine pathway be considered as a next-generation therapeutic target for Parkinson's disease? An update information.Biosci Trends. 2022 Sep 17;16(4):249-256. doi: 10.5582/bst.2022.01352. Epub 2022 Aug 24. Biosci Trends. 2022. PMID: 36002303 Review.
-
Involvement of the kynurenine pathway in the pathogenesis of Parkinson's disease.Prog Neurobiol. 2017 Aug;155:76-95. doi: 10.1016/j.pneurobio.2015.12.009. Epub 2016 Apr 9. Prog Neurobiol. 2017. PMID: 27072742 Review.
Cited by
-
NAD+ metabolism: pathophysiologic mechanisms and therapeutic potential.Signal Transduct Target Ther. 2020 Oct 7;5(1):227. doi: 10.1038/s41392-020-00311-7. Signal Transduct Target Ther. 2020. PMID: 33028824 Free PMC article. Review.
-
NAD+ Metabolism and Diseases with Motor Dysfunction.Genes (Basel). 2021 Nov 9;12(11):1776. doi: 10.3390/genes12111776. Genes (Basel). 2021. PMID: 34828382 Free PMC article. Review.
-
The kynurenine pathway: a finger in every pie.Mol Psychiatry. 2020 Jan;25(1):131-147. doi: 10.1038/s41380-019-0414-4. Epub 2019 Apr 12. Mol Psychiatry. 2020. PMID: 30980044 Free PMC article. Review.
-
C-Reactive protein and the kynurenic acid to quinolinic acid ratio are independently associated with white matter integrity in major depressive disorder.Brain Behav Immun. 2022 Oct;105:180-189. doi: 10.1016/j.bbi.2022.07.011. Epub 2022 Jul 16. Brain Behav Immun. 2022. PMID: 35853557 Free PMC article.
-
Nicotinamide adenine dinucleotide and the sirtuins caution: Pro-cancer functions.Aging Med (Milton). 2021 Nov 30;4(4):337-344. doi: 10.1002/agm2.12184. eCollection 2021 Dec. Aging Med (Milton). 2021. PMID: 34964015 Free PMC article. Review.
References
-
- Parkinson Disease Foundation webpage, Statistics on Parkinson’s Disease, Accessed 20 September, 2017.
-
- Bose A, & Beal MF (2016) Mitochondrial dysfunction in Parkinson’s disease. J Neurochem, 139, 216–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

