Bile acid metabolism and signaling in liver disease and therapy
- PMID: 29104811
- PMCID: PMC5663306
- DOI: 10.1016/j.livres.2017.05.001
Bile acid metabolism and signaling in liver disease and therapy
Abstract
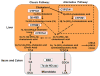
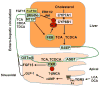
Bile acids play a critical role in the regulation of glucose, lipid, and energy metabolism through activation of the nuclear bile acid receptor farnesoid X receptor (FXR) and membrane G protein-coupled bile acid receptor-1 (Gpbar-1, aka TGR5). Agonist activation of FXR and TGR5 improves insulin and glucose sensitivity and stimulates energy metabolism to prevent diabetes, obesity, and non-alcoholic fatty liver disease (NAFLD). Bile acids have both pro- and anti-inflammatory actions through FXR and TGR5 in the intestine and liver. In the intestine, bile acids activate FXR and TGR5 to stimulate stimulate fibroblast growth factor 15 and glucagon-like peptide-1 secretion. FXR and TGR5 agonists may have therapeutic potential for treating liver-related metabolic diseases, such as diabetes and NAFLD.
Keywords: Bile acid metabolism; Cholestatic liver; diseases Metabolic diseases.
Figures

Similar articles
-
Intestinal Farnesoid X Receptor and Takeda G Protein Couple Receptor 5 Signaling in Metabolic Regulation.Dig Dis. 2017;35(3):241-245. doi: 10.1159/000450981. Epub 2017 Mar 1. Dig Dis. 2017. PMID: 28249273 Free PMC article. Review.
-
Farnesoid X receptor induces Takeda G-protein receptor 5 cross-talk to regulate bile acid synthesis and hepatic metabolism.J Biol Chem. 2017 Jun 30;292(26):11055-11069. doi: 10.1074/jbc.M117.784322. Epub 2017 May 6. J Biol Chem. 2017. PMID: 28478385 Free PMC article.
-
Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism.Hepatology. 2018 Oct;68(4):1574-1588. doi: 10.1002/hep.29857. Epub 2018 May 21. Hepatology. 2018. PMID: 29486523 Free PMC article.
-
Bile acids as regulators of hepatic lipid and glucose metabolism.Dig Dis. 2010;28(1):220-4. doi: 10.1159/000282091. Epub 2010 May 7. Dig Dis. 2010. PMID: 20460915 Review.
-
Crosstalk between FXR and TGR5 controls glucagon-like peptide 1 secretion to maintain glycemic homeostasis.Lab Anim Res. 2018 Dec;34(4):140-146. doi: 10.5625/lar.2018.34.4.140. Epub 2018 Dec 31. Lab Anim Res. 2018. PMID: 30671099 Free PMC article. Review.
Cited by
-
Cross-talk between macrophages and gut microbiota in inflammatory bowel disease: a dynamic interplay influencing pathogenesis and therapy.Front Med (Lausanne). 2024 Sep 16;11:1457218. doi: 10.3389/fmed.2024.1457218. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39355844 Free PMC article. Review.
-
The Role of CYP3A in Health and Disease.Biomedicines. 2022 Oct 24;10(11):2686. doi: 10.3390/biomedicines10112686. Biomedicines. 2022. PMID: 36359206 Free PMC article. Review.
-
Bile Acid and Fibroblast Growth Factor 19 Regulation in Obese Diabetics, and Non-Alcoholic Fatty Liver Disease after Sleeve Gastrectomy.J Clin Med. 2019 Jun 7;8(6):815. doi: 10.3390/jcm8060815. J Clin Med. 2019. PMID: 31181641 Free PMC article.
-
Determination of free and conjugated bile acids in serum of Apoe(-/-) mice fed different lingonberry fractions by UHPLC-MS.Sci Rep. 2019 Mar 7;9(1):3800. doi: 10.1038/s41598-019-40272-8. Sci Rep. 2019. PMID: 30846721 Free PMC article.
-
Gut Microbiota: Novel Therapeutic Target of Ginsenosides for the Treatment of Obesity and Its Complications.Front Pharmacol. 2021 Aug 27;12:731288. doi: 10.3389/fphar.2021.731288. eCollection 2021. Front Pharmacol. 2021. PMID: 34512356 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
