Single-Molecule Interactions of a Monoclonal Anti-DNA Antibody with DNA
- PMID: 29104846
- PMCID: PMC5667910
- DOI: 10.1007/s12668-016-0303-0
Single-Molecule Interactions of a Monoclonal Anti-DNA Antibody with DNA
Abstract
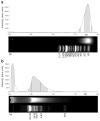
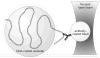
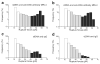
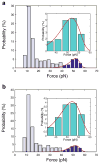
Interactions of DNA with proteins are essential for key biological processes and have both a fundamental and practical significance. In particular, DNA binding to anti-DNA antibodies is a pathogenic mechanism in autoimmune pathology, such as systemic lupus erythematosus. Here we measured at the single-molecule level binding and forced unbinding of surface-attached DNA and a monoclonal anti-DNA antibody MRL4 from a lupus erythematosus mouse. In optical trap-based force spectroscopy, a microscopic antibodycoated latex bead is trapped by a focused laser beam and repeatedly brought into contact with a DNA-coated surface. After careful discrimination of non-specific interactions, we showed that the DNA-antibody rupture force spectra had two regimes, reflecting formation of weaker (20-40 pN) and stronger (>40 pN) immune complexes that implies the existence of at least two bound states with different mechanical stability. The two-dimensional force-free off-rate for the DNA-antibody complexes was ~2.2 × 10-3 s-1, the transition state distance was ~0.94 nm, the apparent on-rate was ~5.26 s-1, and the stiffness of the DNA-antibody complex was characterized by a spring constant of 0.0021 pN/nm, suggesting that the DNA-antibody complex is a relatively stable, but soft and deformable macromolecular structure. The stretching elasticity of the DNA molecules was characteristic of single-stranded DNA, suggesting preferential binding of the MRL4 antibody to one strand of DNA. Collectively, the results provide fundamental characteristics of formation and forced dissociation of DNA-antibody complexes that help to understand principles of DNA-protein interactions and shed light on the molecular basis of autoimmune diseases accompanied by formation of anti-DNA antibodies.
Keywords: Anti-DNA antibody; DNA; Nanomechanics; Optical trap; Single-molecule force spectroscopy; Two-dimensional kinetics.
Figures








References
-
- Williams WM, Isenberg DA. A cross-sectional study of anti-DNA antibodies in the serum and IgG and IgM fraction of healthy individuals, patients with systemic lupus erythematosus and their relatives. Lupus. 1996;5:576–586. - PubMed
-
- Uchida K. Natural antibodies as a sensor of electronegative damage-associated molecular patterns (DAMPs) Free Radical Biology and Medicine. 2014;72:156–161. - PubMed
-
- Lora V, Bonaguri C, Gisondi P, Sandei F, Battistelli L, Russo A, et al. Autoantibody induction and adipokine levels in patients with psoriasis treated with infliximab. Immunologic Research. 2013;56:382–389. - PubMed
-
- Quan CP, Watanabe S, Pamonsinlapatham P, Bouvet J-P. Different dysregulations of the natural antibody repertoire in treated and untreated HIV-1 patients. Journal of Autoimmunity. 2001;17:81–87. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
