Novel Genes Required for the Fitness of Streptococcus pyogenes in Human Saliva
- PMID: 29104937
- PMCID: PMC5663985
- DOI: 10.1128/mSphereDirect.00460-17
Novel Genes Required for the Fitness of Streptococcus pyogenes in Human Saliva
Abstract
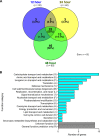
Streptococcus pyogenes (group A streptococcus [GAS]) causes 600 million cases of pharyngitis each year. Despite this considerable disease burden, the molecular mechanisms used by GAS to infect, cause clinical pharyngitis, and persist in the human oropharynx are poorly understood. Saliva is ubiquitous in the human oropharynx and is the first material GAS encounters in the upper respiratory tract. Thus, a fuller understanding of how GAS survives and proliferates in saliva may provide valuable insights into the molecular mechanisms at work in the human oropharynx. We generated a highly saturated transposon insertion mutant library in serotype M1 strain MGAS2221, a strain genetically representative of a pandemic clone that arose in the 1980s and spread globally. The transposon mutant library was exposed to human saliva to screen for GAS genes required for wild-type fitness in this clinically relevant fluid. Using transposon-directed insertion site sequencing (TraDIS), we identified 92 genes required for GAS fitness in saliva. The more prevalent categories represented were genes involved in carbohydrate transport/metabolism, amino acid transport/metabolism, and inorganic ion transport/metabolism. Using six isogenic mutant strains, we confirmed that each of the mutants was significantly impaired for growth or persistence in human saliva ex vivo. Mutants with an inactivated Spy0644 (sptA) or Spy0646 (sptC) gene had especially severe persistence defects. This study is the first to use of TraDIS to study bacterial fitness in human saliva. The new information we obtained will be valuable for future translational maneuvers designed to prevent or treat human GAS infections. IMPORTANCE The human bacterial pathogen Streptococcus pyogenes (group A streptococcus [GAS]) causes more than 600 million cases of pharyngitis annually worldwide, 15 million of which occur in the United States. The human oropharynx is the primary anatomic site for GAS colonization and infection, and saliva is the first material encountered. Using a genome-wide transposon mutant screen, we identified 92 GAS genes required for wild-type fitness in human saliva. Many of the identified genes are involved in carbohydrate transport/metabolism, amino acid transport/metabolism, and inorganic ion transport/metabolism. The new information is potentially valuable for developing novel GAS therapeutics and vaccine research.
Keywords: Streptococcus pyogenes; TraDIS; fitness; human pathogen; saliva; transposon mutagenesis.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
