BioModels: expanding horizons to include more modelling approaches and formats
- PMID: 29106614
- PMCID: PMC5753244
- DOI: 10.1093/nar/gkx1023
BioModels: expanding horizons to include more modelling approaches and formats
Abstract
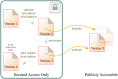
BioModels serves as a central repository of mathematical models representing biological processes. It offers a platform to make mathematical models easily shareable across the systems modelling community, thereby supporting model reuse. To facilitate hosting a broader range of model formats derived from diverse modelling approaches and tools, a new infrastructure for BioModels has been developed that is available at http://www.ebi.ac.uk/biomodels. This new system allows submitting and sharing of a wide range of models with improved support for formats other than SBML. It also offers a version-control backed environment in which authors and curators can work collaboratively to curate models. This article summarises the features available in the current system and discusses the potential benefit they offer to the users over the previous system. In summary, the new portal broadens the scope of models accepted in BioModels and supports collaborative model curation which is crucial for model reproducibility and sharing.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F. et al. . A pathology atlas of the human cancer transcriptome. Science. 2017; 357:6352. - PubMed
-
- Lloyd C.M., Halstead M.D., Nielsen P.F.. CellML: its future, present and past. Prog. Biophys. Mol. Biol. 2004; 85:433–450. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

