Whole genome sequencing of extreme phenotypes identifies variants in CD101 and UBE2V1 associated with increased risk of sexually acquired HIV-1
- PMID: 29108000
- PMCID: PMC5690691
- DOI: 10.1371/journal.ppat.1006703
Whole genome sequencing of extreme phenotypes identifies variants in CD101 and UBE2V1 associated with increased risk of sexually acquired HIV-1
Erratum in
-
Correction: Whole genome sequencing of extreme phenotypes identifies variants in CD101 and UBE2V1 associated with increased risk of sexually acquired HIV-1.PLoS Pathog. 2019 Feb 11;15(2):e1007588. doi: 10.1371/journal.ppat.1007588. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30742678 Free PMC article.
Abstract
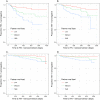
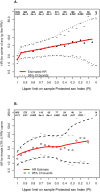
Host genetic variation modifying HIV-1 acquisition risk can inform development of HIV-1 prevention strategies. However, associations between rare or intermediate-frequency variants and HIV-1 acquisition are not well studied. We tested for the association between variation in genic regions and extreme HIV-1 acquisition phenotypes in 100 sub-Saharan Africans with whole genome sequencing data. Missense variants in immunoglobulin-like regions of CD101 and, among women, one missense/5' UTR variant in UBE2V1, were associated with increased HIV-1 acquisition risk (p = 1.9x10-4 and p = 3.7x10-3, respectively, for replication). Both of these genes are known to impact host inflammatory pathways. Effect sizes increased with exposure to HIV-1 after adjusting for the independent effect of increasing exposure on acquisition risk.
Trial registration: ClinicalTrials.gov NCT00194519; NCT00557245.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene Science. 1996;273:1856–62. - PubMed
-
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–77. - PubMed
-
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0 - DOI - PubMed
-
- Fowke KR, Nagelkerke NJ, Kimani J, Simonsen JN, Anzala AO, Bwayo JJ, et al. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet. 1996;348:1347–51. doi: 10.1016/S0140-6736(95)12269-2 - DOI - PubMed
-
- Goh WC, Markee J, Akridge RE, Meldorf M, Musey L, Karchmer T, et al. Protection against human immunodeficiency virus type 1 infection in persons with repeated exposure: evidence for T cell immunity in the absence of inherited CCR5 coreceptor defects. J Infect Dis. 1999;179:548–57. doi: 10.1086/314632 - DOI - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

