Sources of spontaneous mutagenesis in bacteria
- PMID: 29108429
- PMCID: PMC5975382
- DOI: 10.1080/10409238.2017.1394262
Sources of spontaneous mutagenesis in bacteria
Abstract
Mutations in an organism's genome can arise spontaneously, that is, in the absence of exogenous stress and prior to selection. Mutations are often neutral or deleterious to individual fitness but can also provide genetic diversity driving evolution. Mutagenesis in bacteria contributes to the already serious and growing problem of antibiotic resistance. However, the negative impacts of spontaneous mutagenesis on human health are not limited to bacterial antibiotic resistance. Spontaneous mutations also underlie tumorigenesis and evolution of drug resistance. To better understand the causes of genetic change and how they may be manipulated in order to curb antibiotic resistance or the development of cancer, we must acquire a mechanistic understanding of the major sources of mutagenesis. Bacterial systems are particularly well-suited to studying mutagenesis because of their fast growth rate and the panoply of available experimental tools, but efforts to understand mutagenic mechanisms can be complicated by the experimental system employed. Here, we review our current understanding of mutagenic mechanisms in bacteria and describe the methods used to study mutagenesis in bacterial systems.
Keywords: Spontaneous mutagenesis; evolution; fluctuation test; mutation accumulation; mutation reporter; selection.
Conflict of interest statement
Declaration of interest: The authors declare they have no conflict of interest to report.
Figures
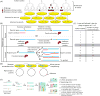

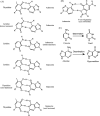
References
-
- Abdulkarim F, Hughes D. Homologous recombination between the tuf genes of Salmonella typhimurium. J Mol Biol. 1996;260:506–22. - PubMed
-
- Banach-Orlowska M, Fijalkowska IJ, Schaaper RM, Jonczyk P. DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol Microbiol. 2005;58:61–70. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
