Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains
- PMID: 29109276
- PMCID: PMC5703309
- DOI: 10.1073/pnas.1712377114
Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains
Abstract
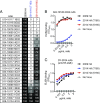
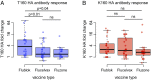
H3N2 viruses continuously acquire mutations in the hemagglutinin (HA) glycoprotein that abrogate binding of human antibodies. During the 2014-2015 influenza season, clade 3C.2a H3N2 viruses possessing a new predicted glycosylation site in antigenic site B of HA emerged, and these viruses remain prevalent today. The 2016-2017 seasonal influenza vaccine was updated to include a clade 3C.2a H3N2 strain; however, the egg-adapted version of this viral strain lacks the new putative glycosylation site. Here, we biochemically demonstrate that the HA antigenic site B of circulating clade 3C.2a viruses is glycosylated. We show that antibodies elicited in ferrets and humans exposed to the egg-adapted 2016-2017 H3N2 vaccine strain poorly neutralize a glycosylated clade 3C.2a H3N2 virus. Importantly, antibodies elicited in ferrets infected with the current circulating H3N2 viral strain (that possesses the glycosylation site) and humans vaccinated with baculovirus-expressed H3 antigens (that possess the glycosylation site motif) were able to efficiently recognize a glycosylated clade 3C.2a H3N2 virus. We propose that differences in glycosylation between H3N2 egg-adapted vaccines and circulating strains likely contributed to reduced vaccine effectiveness during the 2016-2017 influenza season. Furthermore, our data suggest that influenza virus antigens prepared via systems not reliant on egg adaptations are more likely to elicit protective antibody responses that are not affected by glycosylation of antigenic site B of H3N2 HA.
Keywords: antibody; hemagglutinin; influenza; vaccine.
Copyright © 2017 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: S.J.Z., K.P., M.E.G., K.K., S.D.P., P.C.W., A.J.S., S.C., and S.E.H. have no conflicts of interest. J.J.T. is an advisor (nonpaid) for Protein Sciences and is on the scientific advisory board or received consulting payments for Sequiris, Medicago, Takeda, and Flugen. J.J.T.’s laboratory has also received support from Sanofi.
Figures


Comment in
-
Influenza H3N2 Vaccines: Recent Challenges.Trends Microbiol. 2018 Feb;26(2):87-89. doi: 10.1016/j.tim.2017.12.003. Trends Microbiol. 2018. PMID: 29268980
References
-
- Belongia EA, et al. Variable influenza vaccine effectiveness by subtype: A systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis. 2016;16:942–951. - PubMed
-
- Carrat F, Flahault A. Influenza vaccine: The challenge of antigenic drift. Vaccine. 2007;25:6852–6862. - PubMed
-
- Schultz-Cherry S, Jones JC. Influenza vaccines: The good, the bad, and the eggs. Adv Virus Res. 2010;77:63–84. - PubMed
-
- Krammer F, Palese P, Steel J. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr Top Microbiol Immunol. 2015;386:301–321. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

