Anaerobic Oxidation of Ethane, Propane, and Butane by Marine Microbes: A Mini Review
- PMID: 29109712
- PMCID: PMC5660070
- DOI: 10.3389/fmicb.2017.02056
Anaerobic Oxidation of Ethane, Propane, and Butane by Marine Microbes: A Mini Review
Abstract
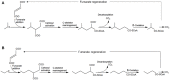
The deep ocean and its sediments are a continuous source of non-methane short-chain alkanes (SCAs) including ethane, propane, and butane. Their high global warming potential, and contribution to local carbon and sulfur budgets has drawn significant scientific attention. Importantly, microbes can use gaseous alkanes and oxidize them to CO2, thus acting as effective biofilters. A relative decrease of these gases with a concomitant 13C enrichment of propane and n-butane in interstitial waters vs. the source suggests microbial anaerobic oxidation. The reported uncoupling of sulfate-reduction (SR) from anaerobic methane oxidation supports their microbial consumption. To date, strain BuS5 isolated from the sediments of Guaymas Basin, Gulf of California, is the only pure culture that can anaerobically degrade propane and n-butane. This organism belongs to a metabolically diverse cluster within the Deltaproteobacteria called Desulfosarcina/Desulfococcus. Other phylotypes involved in gaseous alkane degradation were identified based on stable-isotope labeling and fluorescence in-situ hybridization. A novel syntrophic association of the archaeal genus, Candidatus Syntrophoarchaeum, and a thermophilic SR bacterium, HotSeep-1 was recently discovered from the Guaymas basin, Gulf of California that can anaerobically oxidize n-butane. Strikingly, metagenomic data and the draft genomes of ca. Syntrophoarchaeum suggest that this organism uses a novel mechanism for n-butane oxidation, distinct from the well-established fumarate addition mechanism. These recent findings indicate that a lot remains to be understood about our understanding of anaerobic SCA degradation. This mini-review summarizes our current understanding of microbial anaerobic SCA degradation, and provides an outlook for future research.
Keywords: Desulfosarcina/Desulfococcus; Gulf of Mexico; anaerobic oxidation; short-chain alkanes; sulfate reduction.
Figures

Similar articles
-
The anaerobic degradation of gaseous, nonmethane alkanes - From in situ processes to microorganisms.Comput Struct Biotechnol J. 2015 Mar 19;13:222-8. doi: 10.1016/j.csbj.2015.03.002. eCollection 2015. Comput Struct Biotechnol J. 2015. PMID: 25904994 Free PMC article. Review.
-
Geomicrobiological linkages between short-chain alkane consumption and sulfate reduction rates in seep sediments.Front Microbiol. 2013 Dec 12;4:386. doi: 10.3389/fmicb.2013.00386. eCollection 2013. Front Microbiol. 2013. PMID: 24376442 Free PMC article.
-
Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria.Nature. 2007 Oct 18;449(7164):898-901. doi: 10.1038/nature06200. Epub 2007 Sep 19. Nature. 2007. PMID: 17882164
-
"Candidatus Ethanoperedens," a Thermophilic Genus of Archaea Mediating the Anaerobic Oxidation of Ethane.mBio. 2020 Apr 21;11(2):e00600-20. doi: 10.1128/mBio.00600-20. mBio. 2020. PMID: 32317322 Free PMC article.
-
Anaerobic Degradation of Alkanes by Marine Archaea.Annu Rev Microbiol. 2022 Sep 8;76:553-577. doi: 10.1146/annurev-micro-111021-045911. Epub 2022 Aug 2. Annu Rev Microbiol. 2022. PMID: 35917471 Review.
Cited by
-
Directed Evolution of Protein-Based Sensors for Anaerobic Biological Activation of Methane.Biosensors (Basel). 2024 Jun 30;14(7):325. doi: 10.3390/bios14070325. Biosensors (Basel). 2024. PMID: 39056601 Free PMC article.
-
Variations in microbial community compositions and processes imposed under contrast geochemical contexts in Sicilian mud volcanoes, Italy.Front Microbiol. 2024 Sep 20;15:1461252. doi: 10.3389/fmicb.2024.1461252. eCollection 2024. Front Microbiol. 2024. PMID: 39372275 Free PMC article.
-
Methyl (Alkyl)-Coenzyme M Reductases: Nickel F-430-Containing Enzymes Involved in Anaerobic Methane Formation and in Anaerobic Oxidation of Methane or of Short Chain Alkanes.Biochemistry. 2019 Dec 31;58(52):5198-5220. doi: 10.1021/acs.biochem.9b00164. Epub 2019 Apr 5. Biochemistry. 2019. PMID: 30951290 Free PMC article. Review.
-
Anaerobic oxidation of ammonium and short-chain gaseous alkanes coupled to nitrate reduction by a bacterial consortium.ISME J. 2024 Jan 8;18(1):wrae063. doi: 10.1093/ismejo/wrae063. ISME J. 2024. PMID: 38624180 Free PMC article.
-
In Situ Metabolic Rates of Alkane-Degrading Sulphate-Reducing Bacteria in Hydrocarbon Seep Sediments Revealed by Combining CARD-FISH, NanoSIMS, and Mathematical Modelling.Environ Microbiol. 2025 Aug;27(8):e70151. doi: 10.1111/1462-2920.70151. Environ Microbiol. 2025. PMID: 40728192 Free PMC article.
References
-
- Aitken C. M., Jones D. M., Maguire M. J., Gray N. D., Sherry A., Bowler B. F. J., et al. (2013). Evidence that crude oil alkane activation proceeds by different mechanisms under sulfate-reducing and methanogenic conditions. Geochim. Cosmochim. Acta 109, 162–174. 10.1016/j.gca.2013.01.031 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

