Structure-Affinity Relationships and Structure-Kinetic Relationships of 1,2-Diarylimidazol-4-carboxamide Derivatives as Human Cannabinoid 1 Receptor Antagonists
- PMID: 29111736
- PMCID: PMC5734604
- DOI: 10.1021/acs.jmedchem.7b00861
Structure-Affinity Relationships and Structure-Kinetic Relationships of 1,2-Diarylimidazol-4-carboxamide Derivatives as Human Cannabinoid 1 Receptor Antagonists
Abstract
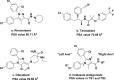
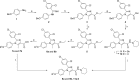
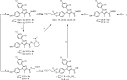
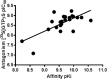
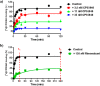
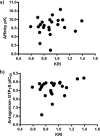
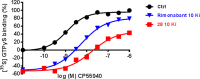
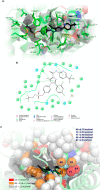
We report on the synthesis and biological evaluation of a series of 1,2-diarylimidazol-4-carboxamide derivatives developed as CB1 receptor antagonists. These were evaluated in a radioligand displacement binding assay, a [35S]GTPγS binding assay, and in a competition association assay that enables the relatively fast kinetic screening of multiple compounds. The compounds show high affinities and a diverse range of kinetic profiles at the CB1 receptor and their structure-kinetic relationships (SKRs) were established. Using the recently resolved hCB1 receptor crystal structures, we also performed a modeling study that sheds light on the crucial interactions for both the affinity and dissociation kinetics of this family of ligands. We provide evidence that, next to affinity, additional knowledge of binding kinetics is useful for selecting new hCB1 receptor antagonists in the early phases of drug discovery.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Kinetics of human cannabinoid 1 (CB1) receptor antagonists: Structure-kinetics relationships (SKR) and implications for insurmountable antagonism.Biochem Pharmacol. 2018 May;151:166-179. doi: 10.1016/j.bcp.2017.10.014. Epub 2017 Nov 2. Biochem Pharmacol. 2018. PMID: 29102677
-
Synthesis and activity of 1,3,5-triphenylimidazolidine-2,4-diones and 1,3,5-triphenyl-2-thioxoimidazolidin-4-ones: characterization of new CB1 cannabinoid receptor inverse agonists/antagonists.J Med Chem. 2006 Feb 9;49(3):872-82. doi: 10.1021/jm050484f. J Med Chem. 2006. PMID: 16451053
-
Structure-Affinity Relationships and Structure-Kinetics Relationships of Pyrido[2,1-f]purine-2,4-dione Derivatives as Human Adenosine A3 Receptor Antagonists.J Med Chem. 2017 Sep 14;60(17):7555-7568. doi: 10.1021/acs.jmedchem.7b00950. Epub 2017 Aug 30. J Med Chem. 2017. PMID: 28806076 Free PMC article.
-
Bioisosteric replacements of the pyrazole moiety of rimonabant: synthesis, biological properties, and molecular modeling investigations of thiazoles, triazoles, and imidazoles as potent and selective CB1 cannabinoid receptor antagonists.J Med Chem. 2005 Mar 24;48(6):1823-38. doi: 10.1021/jm040843r. J Med Chem. 2005. PMID: 15771428
-
Insight into Thermodynamic and Kinetic Profiles in Small-Molecule Optimization.J Med Chem. 2022 Aug 25;65(16):10809-10847. doi: 10.1021/acs.jmedchem.2c00682. Epub 2022 Aug 15. J Med Chem. 2022. PMID: 35969687 Review.
Cited by
-
Binding kinetics of ligands acting at GPCRs.Mol Cell Endocrinol. 2019 Apr 5;485:9-19. doi: 10.1016/j.mce.2019.01.018. Epub 2019 Feb 8. Mol Cell Endocrinol. 2019. PMID: 30738950 Free PMC article. Review.
-
Perspective: Implications of Ligand-Receptor Binding Kinetics for Therapeutic Targeting of G Protein-Coupled Receptors.ACS Pharmacol Transl Sci. 2020 Mar 18;3(2):179-189. doi: 10.1021/acsptsci.0c00012. eCollection 2020 Apr 10. ACS Pharmacol Transl Sci. 2020. PMID: 32296761 Free PMC article. Review.
-
A universal cannabinoid CB1 and CB2 receptor TR-FRET kinetic ligand-binding assay.Front Pharmacol. 2025 Apr 9;16:1469986. doi: 10.3389/fphar.2025.1469986. eCollection 2025. Front Pharmacol. 2025. PMID: 40271066 Free PMC article.
References
-
- Pertwee R. G.; Howlett A. C.; Abood M. E.; Alexander S. P. H.; Di Marzo V.; Elphick M. R.; Greasley P. J.; Hansen H. S.; Kunos G.; Mackie K.; Mechoulam R.; Ross R. A. International union of basic and clinical pharmacology. LXXIX. Cannabinoid receptors and their ligands: beyond CB1 and CB2. Pharmacol. Rev. 2010, 62, 588–631. 10.1124/pr.110.003004. - DOI - PMC - PubMed
-
- Abood M.; Barth F.; Bonner T. I.; Cabral G.; Casellas P.; Cravatt B. F.; Devane W. A.; Elphick M. R.; Felder C. C.; Herkenham M.; Howlett A. C.; Kunos G.; Mackie K.; Mechoulam R.; Pertwee R. G.. Cannabinoid receptors: CB1 receptor. In IUPHAR/BPS Guide to Pharmacology; International Union of Basic and Clinical Pharmacology, May 16, 2017; http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=56 (accessed July 20, 2017).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

