Dual-Located WHIRLY1 Interacting with LHCA1 Alters Photochemical Activities of Photosystem I and Is Involved in Light Adaptation in Arabidopsis
- PMID: 29112140
- PMCID: PMC5713321
- DOI: 10.3390/ijms18112352
Dual-Located WHIRLY1 Interacting with LHCA1 Alters Photochemical Activities of Photosystem I and Is Involved in Light Adaptation in Arabidopsis
Abstract
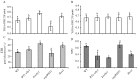
Plastid-nucleus-located WHIRLY1 protein plays a role in regulating leaf senescence and is believed to associate with the increase of reactive oxygen species delivered from redox state of the photosynthetic electron transport chain. In order to make sure whether WHIRLY1 plays a role in photosynthesis, in this study, the performances of photosynthesis were detected in Arabidopsis whirly1 knockout (kowhy1) and plastid localized WHIRLY1 overexpression (oepWHY1) plants. Loss of WHIRLY1 leads to a higher photochemical quantum yield of photosystem I Y(I) and electron transport rate (ETR) and a lower non-photochemical quenching (NPQ) involved in the thermal dissipation of excitation energy of chlorophyll fluorescence than the wild type. Further analyses showed that WHIRLY1 interacts with Light-harvesting protein complex I (LHCA1) and affects the expression of genes encoding photosystem I (PSI) and light harvest complexes (LHCI). Moreover, loss of WHIRLY1 decreases chloroplast NAD(P)H dehydrogenase-like complex (NDH) activity and the accumulation of NDH supercomplex. Several genes encoding the PSI-NDH complexes are also up-regulated in kowhy1 and the whirly1whirly3 double mutant (ko1/3) but steady in oepWHY1 plants. However, under high light conditions (800 μmol m-2 s-1), both kowhy1 and ko1/3 plants show lower ETR than wild-type which are contrary to that under normal light condition. Moreover, the expression of several PSI-NDH encoding genes and ERF109 which is related to jasmonate (JA) response varied in kowhy1 under different light conditions. These results indicate that WHIRLY1 is involved in the alteration of ETR by affecting the activities of PSI and supercomplex formation of PSI with LHCI or NDH and may acting as a communicator between the plastids and the nucleus.
Keywords: WHIRLY1; electron transport rate (ETR); light; photochemical activities; photosystem I; plastid gene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





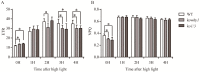

References
-
- Pfannschmidt T., Nilsson A., Allen J.F. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

