Poxvirus Host Range Genes and Virus-Host Spectrum: A Critical Review
- PMID: 29112165
- PMCID: PMC5707538
- DOI: 10.3390/v9110331
Poxvirus Host Range Genes and Virus-Host Spectrum: A Critical Review
Abstract
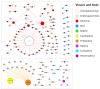
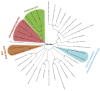
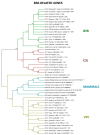
The Poxviridae family is comprised of double-stranded DNA viruses belonging to nucleocytoplasmic large DNA viruses (NCLDV). Among the NCLDV, poxviruses exhibit the widest known host range, which is likely observed because this viral family has been more heavily investigated. However, relative to each member of the Poxviridae family, the spectrum of the host is variable, where certain viruses can infect a large range of hosts, while others are restricted to only one host species. It has been suggested that the variability in host spectrum among poxviruses is linked with the presence or absence of some host range genes. Would it be possible to extrapolate the restriction of viral replication in a specific cell lineage to an animal, a far more complex organism? In this study, we compare and discuss the relationship between the host range of poxvirus species and the abundance/diversity of host range genes. We analyzed the sequences of 38 previously identified and putative homologs of poxvirus host range genes, and updated these data with deposited sequences of new poxvirus genomes. Overall, the term host range genes might not be the most appropriate for these genes, since no correlation between them and the viruses' host spectrum was observed, and a change in nomenclature should be considered. Finally, we analyzed the evolutionary history of these genes, and reaffirmed the occurrence of horizontal gene transfer (HGT) for certain elements, as previously suggested. Considering the data presented in this study, it is not possible to associate the diversity of host range factors with the amount of hosts of known poxviruses, and this traditional nomenclature creates misunderstandings.
Keywords: Poxviridae; evolution; horizontal gene transfer; host range genes; network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- International Committee on Taxonomy of Viruses—Taxonomy. [(accessed on 1 July 2017)]; Available online: https://talk.ictvonline.org/taxonomy/w/ictv-taxonomy.
-
- Fenner F., Henderson D.A., Arita I., Jezek Z., Ladnyi I.D. Smallpox and Its Eradication. 1st ed. World Health Organization; Geneva, Switzerland: 1988. pp. 1371–1409.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

