VarCards: an integrated genetic and clinical database for coding variants in the human genome
- PMID: 29112736
- PMCID: PMC5753295
- DOI: 10.1093/nar/gkx1039
VarCards: an integrated genetic and clinical database for coding variants in the human genome
Abstract
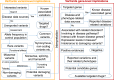
A growing number of genomic tools and databases were developed to facilitate the interpretation of genomic variants, particularly in coding regions. However, these tools are separately available in different online websites or databases, making it challenging for general clinicians, geneticists and biologists to obtain the first-hand information regarding some particular variants and genes of interest. Starting with coding regions and splice sties, we artificially generated all possible single nucleotide variants (n = 110 154 363) and cataloged all reported insertion and deletions (n = 1 223 370). We then annotated these variants with respect to functional consequences from more than 60 genomic data sources to develop a database, named VarCards (http://varcards.biols.ac.cn/), by which users can conveniently search, browse and annotate the variant- and gene-level implications of given variants, including the following information: (i) functional effects; (ii) functional consequences through different in silico algorithms; (iii) allele frequencies in different populations; (iv) disease- and phenotype-related knowledge; (v) general meaningful gene-level information; and (vi) drug-gene interactions. As a case study, we successfully employed VarCards in interpretation of de novo mutations in autism spectrum disorders. In conclusion, VarCards provides an intuitive interface of necessary information for researchers to prioritize candidate variations and genes.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Rabbani B., Tekin M., Mahdieh N.. The promise of whole-exome sequencing in medical genetics. J. Hum. Genet. 2013; 59:5–15. - PubMed
-
- Biesecker L.G., Green R.C.. Diagnostic clinical genome and exome sequencing. N. Engl. J. Med. 2014; 370:2418–2425. - PubMed
-
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E. et al. . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015; 17:405–423. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

