Long-Fiber Carbon Nanotubes Replicate Asbestos-Induced Mesothelioma with Disruption of the Tumor Suppressor Gene Cdkn2a (Ink4a/Arf)
- PMID: 29112861
- PMCID: PMC5681354
- DOI: 10.1016/j.cub.2017.09.007
Long-Fiber Carbon Nanotubes Replicate Asbestos-Induced Mesothelioma with Disruption of the Tumor Suppressor Gene Cdkn2a (Ink4a/Arf)
Abstract
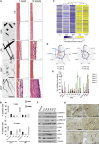
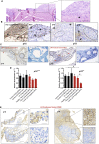
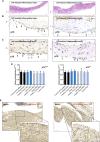
Mesothelioma is a fatal tumor of the pleura and is strongly associated with asbestos exposure. The molecular mechanisms underlying the long latency period of mesothelioma and driving carcinogenesis are unknown. Moreover, late diagnosis means that mesothelioma research is commonly focused on end-stage disease. Although disruption of the CDKN2A (INK4A/ARF) locus has been reported in end-stage disease, information is lacking on the status of this key tumor suppressor gene in pleural lesions preceding mesothelioma. Manufactured carbon nanotubes (CNTs) are similar to asbestos in terms of their fibrous shape and biopersistent properties and thus may pose an asbestos-like inhalation hazard. Here we show that instillation of either long CNTs or long asbestos fibers into the pleural cavity of mice induces mesothelioma that exhibits common key pro-oncogenic molecular events throughout the latency period of disease progression. Sustained activation of pro-oncogenic signaling pathways, increased proliferation, and oxidative DNA damage form a common molecular signature of long-CNT- and long-asbestos-fiber-induced pathology. We show that hypermethylation of p16/Ink4a and p19/Arf in CNT- and asbestos-induced inflammatory lesions precedes mesothelioma; this results in silencing of Cdkn2a (Ink4a/Arf) and loss of p16 and p19 protein, consistent with epigenetic alterations playing a gatekeeper role in cancer. In end-stage mesothelioma, silencing of p16/Ink4a is sustained and deletion of p19/Arf is detected, recapitulating human disease. This study addresses the long-standing question of which early molecular changes drive carcinogenesis during the long latency period of mesothelioma development and shows that CNT and asbestos pose a similar health hazard.
Keywords: CDKN2A; asbestos; carbon nanotubes; epigenetics; hypermethylation; mesothelioma; toxicity; tumor suppressor genes.
Copyright © 2017 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures




Comment in
-
Mesothelioma: Identical Routes to Malignancy from Asbestos and Carbon Nanotubes.Curr Biol. 2017 Nov 6;27(21):R1173-R1176. doi: 10.1016/j.cub.2017.07.026. Curr Biol. 2017. PMID: 29112873
References
-
- Murphy F.A., Poland C.A., Duffin R., Al-Jamal K.T., Ali-Boucetta H., Nunes A., Byrne F., Prina-Mello A., Volkov Y., Li S. Length-dependent retention of carbon nanotubes in the pleural space of mice initiates sustained inflammation and progressive fibrosis on the parietal pleura. Am. J. Pathol. 2011;178:2587–2600. - PMC - PubMed
-
- Nagai H., Toyokuni S. Biopersistent fiber-induced inflammation and carcinogenesis: lessons learned from asbestos toward safety of fibrous nanomaterials. Arch. Biochem. Biophys. 2010;502:1–7. - PubMed
-
- Lecomte C., Andujar P., Renier A., Kheuang L., Abramowski V., Mellottee L., Fleury-Feith J., Zucman-Rossi J., Giovannini M., Jaurand M.C. Similar tumor suppressor gene alteration profiles in asbestos-induced murine and human mesothelioma. Cell Cycle. 2005;4:1862–1869. - PubMed
-
- Guo G., Chmielecki J., Goparaju C., Heguy A., Dolgalev I., Carbone M., Seepo S., Meyerson M., Pass H.I. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 2015;75:264–269. - PubMed
-
- Lo Iacono M., Monica V., Righi L., Grosso F., Libener R., Vatrano S., Bironzo P., Novello S., Musmeci L., Volante M. Targeted next-generation sequencing of cancer genes in advanced stage malignant pleural mesothelioma: a retrospective study. J. Thorac. Oncol. 2015;10:492–499. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

