Natural product β-thujaplicin inhibits homologous recombination repair and sensitizes cancer cells to radiation therapy
- PMID: 29112893
- PMCID: PMC5909195
- DOI: 10.1016/j.dnarep.2017.10.009
Natural product β-thujaplicin inhibits homologous recombination repair and sensitizes cancer cells to radiation therapy
Abstract
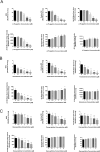
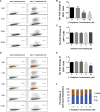
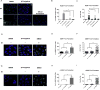
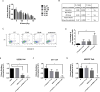
Investigation of natural products is an attractive strategy to identify novel compounds for cancer prevention and treatment. Numerous studies have shown the efficacy and safety of natural products, and they have been widely used as alternative treatments for a wide range of illnesses, including cancers. However, it remains unknown whether natural products affect homologous recombination (HR)-mediated DNA repair and whether these compounds can be used as sensitizers with minimal toxicity to improve patients' responses to radiation therapy, a mainstay of treatment for many human cancers. In this study, in order to systematically identify natural products with an inhibitory effect on HR repair, we developed a high-throughput image-based HR repair screening assay and screened a chemical library containing natural products. Among the most interesting of the candidate compounds identified from the screen was β-thujaplicin, a bioactive compound isolated from the heart wood of plants in the Cupressaceae family, can significantly inhibit HR repair. We further demonstrated that β-thujaplicin inhibits HR repair by reducing the recruitment of a key HR repair protein, Rad51, to DNA double-strand breaks. More importantly, our results showed that β-thujaplicin can radiosensitize cancer cells. Additionally, β-thujaplicin sensitizes cancer cells to PARP inhibitor in different cancer cell lines. Collectively, our findings for the first time identify natural compound β-thujaplicin, which has a good biosafety profile, as a novel HR repair inhibitor with great potential to be translated into clinical applications as a sensitizer to DNA-damage-inducing treatment such as radiation and PARP inhibitor. In addition, our study provides proof of the principle that our robust high-throughput functional HR repair assay can be used for a large-scale screening system to identify novel natural products that regulate DNA repair and cellular responses to DNA damage-inducing treatments such as radiation therapy.
Keywords: DNA repair; Homologous recombination; PARP inhibitor; Radiosensitizer; β-thujaplicin.
Copyright © 2017 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics 2014. CA: Cancer J Clin. 2014;64:83–103. - PubMed
-
- Schwarz R, Bruland O, Cassoni A, Schomberg P, Bielack S. The role of radiotherapy in oseosarcoma. Cancer Treat Res. 2009;152:147–164. - PubMed
-
- Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treatment Rev. 2014;40:523–532. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

