Karyotype relationships among selected deer species and cattle revealed by bovine FISH probes
- PMID: 29112970
- PMCID: PMC5675437
- DOI: 10.1371/journal.pone.0187559
Karyotype relationships among selected deer species and cattle revealed by bovine FISH probes
Abstract
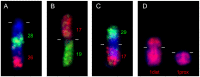
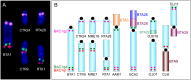
The Cervidae family comprises more than fifty species divided into three subfamilies: Capreolinae, Cervinae and Hydropotinae. A characteristic attribute for the species included in this family is the great karyotype diversity, with the chromosomal numbers ranging from 2n = 6 observed in female Muntiacus muntjak vaginalis to 2n = 70 found in Mazama gouazoubira as a result of numerous Robertsonian and tandem fusions. This work reports chromosomal homologies between cattle (Bos taurus, 2n = 60) and nine cervid species using a combination of whole chromosome and region-specific paints and BAC clones derived from cattle. We show that despite the great diversity of karyotypes in the studied species, the number of conserved chromosomal segments detected by 29 cattle whole chromosome painting probes was 35 for all Cervidae samples. The detailed analysis of the X chromosomes revealed two different morphological types within Cervidae. The first one, present in the Capreolinae is a sub/metacentric X with the structure more similar to the bovine X. The second type found in Cervini and Muntiacini is an acrocentric X which shows rearrangements in the proximal part that have not yet been identified within Ruminantia. Moreover, we characterised four repetitive sequences organized in heterochromatic blocks on sex chromosomes of the reindeer (Rangifer tarandus). We show that these repeats gave no hybridization signals to the chromosomes of the closely related moose (Alces alces) and are therefore specific to the reindeer.
Conflict of interest statement
Figures





References
-
- Wilson DE, Reeder DM. Mammal Species of the World. Johns Hopkins University Press, Baltimore; 2005.
-
- Fontana F, Rubini M. Chromosomal evolution in Cervidae. BioSystems. 1990;24: 157–174. - PubMed
-
- Bonnet-Garnier A, Claro F, Thévenon S, Gautier M, Hayes H. Identification by R-banding and FISH of chromosome arms involved in Robertsonian translocations in several deer species. Chromosome Res. 2003;11: 649–663. - PubMed
-
- Chi JX, Huang L, Nie W, Wang J, Su B, Yang F. Defining the orientation of the tandem fusions that occurred during the evolution of Indian muntjac chromosomes by BAC mapping. Chromosoma. 2005;114: 167–172. doi: 10.1007/s00412-005-0004-x - DOI - PubMed
-
- Yang F, O’Brien PC, Wienberg J, Ferguson-Smith MA. A reappraisal of the tandem fusion theory of karyotype evolution in Indian muntjac using chromosome painting. Chromosome Res. 1997;5: 109–117. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

