A generic Transcriptomics Reporting Framework (TRF) for 'omics data processing and analysis
- PMID: 29113939
- PMCID: PMC6826341
- DOI: 10.1016/j.yrtph.2017.11.001
A generic Transcriptomics Reporting Framework (TRF) for 'omics data processing and analysis
Abstract
A generic Transcriptomics Reporting Framework (TRF) is presented that lists parameters that should be reported in 'omics studies used in a regulatory context. The TRF encompasses the processes from transcriptome profiling from data generation to a processed list of differentially expressed genes (DEGs) ready for interpretation. Included within the TRF is a reference baseline analysis (RBA) that encompasses raw data selection; data normalisation; recognition of outliers; and statistical analysis. The TRF itself does not dictate the methodology for data processing, but deals with what should be reported. Its principles are also applicable to sequencing data and other 'omics. In contrast, the RBA specifies a simple data processing and analysis methodology that is designed to provide a comparison point for other approaches and is exemplified here by a case study. By providing transparency on the steps applied during 'omics data processing and analysis, the TRF will increase confidence processing of 'omics data, and regulatory use. Applicability of the TRF is ensured by its simplicity and generality. The TRF can be applied to all types of regulatory 'omics studies, and it can be executed using different commonly available software tools.
Keywords: Bioinformatics; Differentially expressed genes; Gene expression; Normalisation of ‘omics data; Regulatory toxicology; Reproducibility; Statistical analysis.
Crown Copyright © 2017. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest
UGS was hired by ECETOC and the CEFIC LRI to assist in the preparation of the manuscript. The other authors were engaged in the course of their normal employment. The authors alone are responsible for the content and writing of the paper.
Figures

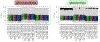

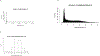
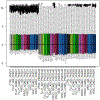


References
-
- Affymetrix, 2013. Affymetrix® GeneChip® Command Console® (AGCC) 4.0 User Manual, available at: http://www.affymetrix.com/support/technical/byproduct.affx?product=comma....
-
- Affymetrix, 2014. Transcriptome Analysis Console (TAC) 3.0. User Guide, available at: http://www.affymetrix.com/estore/browse/level_seven_software_products_on...
-
- Agilent, 2014. Agilent GeneSpring. User manual, available at: http://www.agilent.com/cs/library/usermanuals/public/GeneSpring-manual.pdf.
-
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M, 2001. Minimum information about a microarray experiment (MIAME) - toward standards for microarray data. Nat. Genet 29, 365–371. - PubMed
-
- Buesen R, Chorley BN, da Silva Lima B, Daston G, Deferme L, Ebbels T, Gant TW, Goetz A, Greally J, Gribaldo L, Hackermüller J, Hubesch B, Jennen D, Johnson K, Kanno J, Kauffmann H-M, Laffont M, Meehan R, Pemberton M, Perdichizzi S, Piersma AH, Sauer UG, Schmidt K, Seitz H, Sumida K, Tollefsen KE, Tong W, Tralau T, van Ravenzwaay B, Weber R, Worth A, Yauk C, Poole A, 2017. Applying ‘omics technologies in chemicals risk assessment: Report of an ECETOC workshop Regulat. Toxicol. Pharmacol. epub ahead of print 25 September 2017, doi: 10.1016/j.yrtph.2017.09.002. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

