A Role for the F-Box Protein HAWAIIAN SKIRT in Plant microRNA Function
- PMID: 29114080
- PMCID: PMC5761791
- DOI: 10.1104/pp.17.01313
A Role for the F-Box Protein HAWAIIAN SKIRT in Plant microRNA Function
Abstract
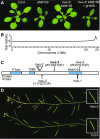
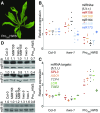
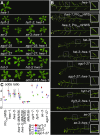
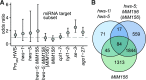
As regulators of gene expression in multicellular organisms, microRNAs (miRNAs) are crucial for growth and development. Although a plethora of factors involved in their biogenesis and action in Arabidopsis (Arabidopsis thaliana) has been described, these processes and their fine-tuning are not fully understood. Here, we used plants expressing an artificial miRNA target mimic (MIM) to screen for negative regulators of miR156. We identified a new mutant allele of the F-box gene HAWAIIAN SKIRT (HWS; At3G61590), hws-5, as a suppressor of the MIM156-induced developmental and molecular phenotypes. In hws plants, levels of some endogenous miRNAs are increased and their mRNA targets decreased. Plants constitutively expressing full-length HWS-but not a truncated version lacking the F-box domain-display morphological and molecular phenotypes resembling those of mutants defective in miRNA biogenesis and activity. In combination with such mutants, hws loses its delayed floral organ abscission ("skirt") phenotype, suggesting epistasis. Also, the hws transcriptome profile partially resembles those of well-known miRNA mutants hyl1-2, se-3, and ago1-27, pointing to a role in a common pathway. We thus propose HWS as a novel, F-box dependent factor involved in miRNA function.
© 2018 American Society of Plant Biologists. All Rights Reserved.
Figures

References
-
- Bak RO, Mikkelsen JG (2014) miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA 5: 317–333 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

