A T7 RNA Polymerase Mutant Enhances the Yield of 5'-Thienoguanosine-Initiated RNAs
- PMID: 29115013
- PMCID: PMC6047071
- DOI: 10.1002/cbic.201700538
A T7 RNA Polymerase Mutant Enhances the Yield of 5'-Thienoguanosine-Initiated RNAs
Abstract
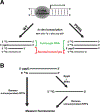
Spectroscopic methods, which are used to establish RNA structure-function relationships, require strategies for post-synthetic, site-specific incorporation of chemical probes into target RNAs. For RNAs larger than 50 nt, the enzymatic incorporation of a nucleoside or nucleotide monophosphate guanosine analogue (G analogue) at their 5'-end is routinely achieved by T7 RNA polymerase (T7RNAP)-mediated in vitro transcription (IVT) of the appropriate DNA template containing a GTP-initiating class III Φ6.5 promoter. However, when high G analogue:GTP ratios are used to bias G analogue incorporation at the 5'-end, RNA yield is compromised. Here, we show that the use of a T7RNAP P266L mutant in IVT with 10:1 thienoguanosine (th G):GTP increased the percent incorporation and yield of 5'-th G-initiated precursor tRNA for a net ≈threefold gain compared to IVT with wild-type T7RNAP. We also demonstrated that a one-pot multienzyme approach, consisting of transcription by T7RNAP P266L and post-transcriptional cleanup by polyphosphatase and an exonuclease, led to essentially near-homogeneous 5'-th G-modified transcripts. This approach should be of broad utility in preparing 5'-modified RNAs.
Keywords: RNA; T7 RNA polymerase; fluorescent probes; in vitro transcription; thienoguanosine.
© 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

References
-
- Alexander S, Devaraj N, Biochemistry 2017, 56, 5185–5193. - PubMed
-
- Paredes E, Evans M, Das SR, Methods 2011, 54, 251–259. - PubMed
-
- a) Lee GH, Lim HK, Jung W, Hah SS, Bull. Korean Chem. Soc 2012, 33, 3861–3863;
- b) Skipsey M, Hack G, Hoope TA, Shankey MC, Conway LP, Schroder M, Hodgson DRW, Nucleos. Nucleot. Nucl 2013, 32, 670–681; - PMC - PubMed
- c) Zhang L, Sun L, Cui Z, Gottlieb RL, Zhang B, Bioconjug. Chem. 2001, 12, 939–948; - PubMed
- d) Williamson D, Cann MJ, Hodgson DRW, Chem Commun. 2007, 47, 5096–5098. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

