Guanylate-binding protein-1 is a potential new therapeutic target for triple-negative breast cancer
- PMID: 29115931
- PMCID: PMC5688804
- DOI: 10.1186/s12885-017-3726-2
Guanylate-binding protein-1 is a potential new therapeutic target for triple-negative breast cancer
Abstract
Background: Triple-negative breast cancer (TNBC) is characterized by a lack of estrogen and progesterone receptor expression (ESR and PGR, respectively) and an absence of human epithelial growth factor receptor (ERBB2) amplification. Approximately 15-20% of breast malignancies are TNBC. Patients with TNBC often have an unfavorable prognosis. In addition, TNBC represents an important clinical challenge since it does not respond to hormone therapy.
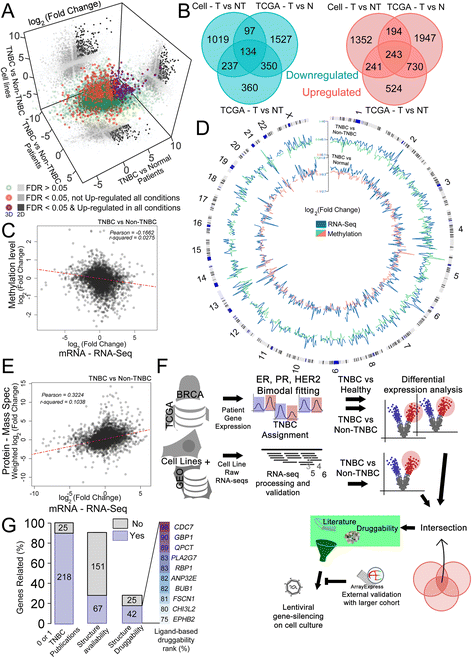
Methods: In this work, we integrated high-throughput mRNA sequencing (RNA-Seq) data from normal and tumor tissues (obtained from The Cancer Genome Atlas, TCGA) and cell lines obtained through in-house sequencing or available from the Gene Expression Omnibus (GEO) to generate a unified list of differentially expressed (DE) genes. Methylome and proteomic data were integrated to our analysis to give further support to our findings. Genes that were overexpressed in TNBC were then curated to retain new potentially druggable targets based on in silico analysis. Knocking-down was used to assess gene importance for TNBC cell proliferation.
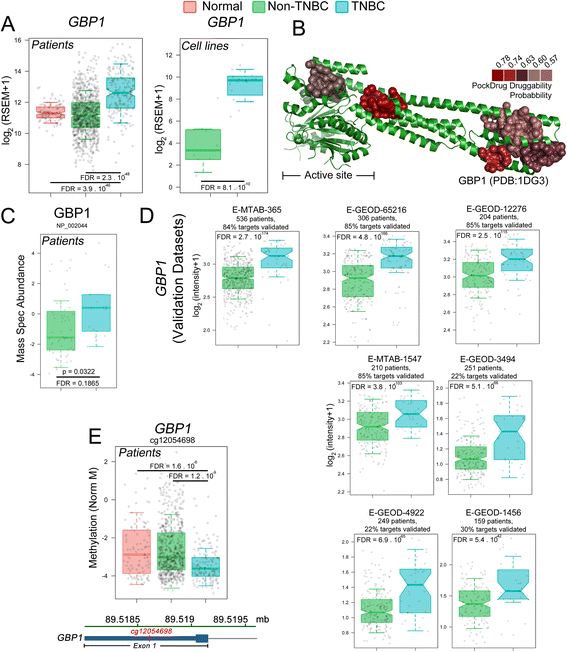
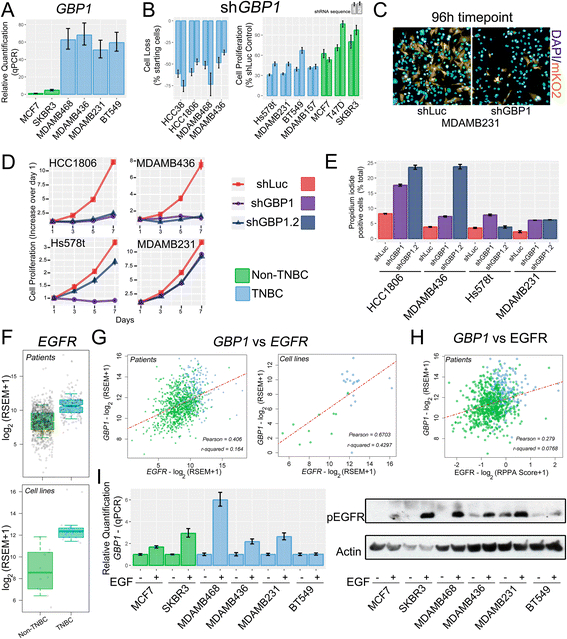
Results: Our pipeline analysis generated a list of 243 potential new targets for treating TNBC. We finally demonstrated that knock-down of Guanylate-Binding Protein 1 (GBP1 ), one of the candidate genes, selectively affected the growth of TNBC cell lines. Moreover, we showed that GBP1 expression was controlled by epidermal growth factor receptor (EGFR) in breast cancer cell lines.
Conclusions: We propose that GBP1 is a new potential druggable therapeutic target for treating TNBC with enhanced EGFR expression.
Keywords: Breast cancer; Gene expression; RNA-Seq; Therapeutic target; Transcriptomics; Triple-negative breast cancer.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
MET is a potential target for use in combination therapy with EGFR inhibition in triple-negative/basal-like breast cancer.Int J Cancer. 2014 May 15;134(10):2424-36. doi: 10.1002/ijc.28566. Int J Cancer. 2014. PMID: 24615768
-
Identification of differentially expressed genes between triple and non-triple-negative breast cancer using bioinformatics analysis.Breast Cancer. 2019 Nov;26(6):784-791. doi: 10.1007/s12282-019-00988-x. Epub 2019 Jun 13. Breast Cancer. 2019. PMID: 31197620
-
Higher levels of TIMP-1 expression are associated with a poor prognosis in triple-negative breast cancer.Mol Cancer. 2016 Apr 30;15(1):30. doi: 10.1186/s12943-016-0515-5. Mol Cancer. 2016. PMID: 27130446 Free PMC article.
-
High-throughput «Omics» technologies: New tools for the study of triple-negative breast cancer.Cancer Lett. 2016 Nov 1;382(1):77-85. doi: 10.1016/j.canlet.2016.03.001. Epub 2016 Mar 7. Cancer Lett. 2016. PMID: 26965997 Review.
-
Perspectives on Epidermal Growth Factor Receptor Regulation in Triple-Negative Breast Cancer: Ligand-Mediated Mechanisms of Receptor Regulation and Potential for Clinical Targeting.Adv Cancer Res. 2015;127:253-81. doi: 10.1016/bs.acr.2015.04.008. Epub 2015 May 8. Adv Cancer Res. 2015. PMID: 26093903 Review.
Cited by
-
Unraveling the Role of Guanylate-Binding Proteins (GBPs) in Breast Cancer: A Comprehensive Literature Review and New Data on Prognosis in Breast Cancer Subtypes.Cancers (Basel). 2022 Jun 4;14(11):2794. doi: 10.3390/cancers14112794. Cancers (Basel). 2022. PMID: 35681772 Free PMC article.
-
Dual inhibition of glutaminase and carnitine palmitoyltransferase decreases growth and migration of glutaminase inhibition-resistant triple-negative breast cancer cells.J Biol Chem. 2019 Jun 14;294(24):9342-9357. doi: 10.1074/jbc.RA119.008180. Epub 2019 Apr 30. J Biol Chem. 2019. PMID: 31040181 Free PMC article.
-
Biological Functions and Identification of Novel Biomarker Expressed on the Surface of Breast Cancer-Derived Cancer Stem Cells via Proteomic Analysis.Mol Cells. 2020 Apr 30;43(4):384-396. doi: 10.14348/molcells.2020.2230. Mol Cells. 2020. PMID: 32235022 Free PMC article.
-
Correlation between the risk of lymph node metastasis and the expression of GBP1 in breast cancer patients.Pak J Med Sci. 2024 Jan-Feb;40(1Part-I):159-164. doi: 10.12669/pjms.40.1.8251. Pak J Med Sci. 2024. PMID: 38196488 Free PMC article.
-
GBP5 Serves as a Potential Marker to Predict a Favorable Response in Triple-Negative Breast Cancer Patients Receiving a Taxane-Based Chemotherapy.J Pers Med. 2021 Mar 12;11(3):197. doi: 10.3390/jpm11030197. J Pers Med. 2021. PMID: 33809079 Free PMC article.
References
-
- Isakov O, Shomron N. Deep Sequencing Data Analysis : Challenges and Solutions. Bioinforma. - Trends Methodol. 2011. p. 655–679.
-
- Mardis ER. The impact of next-generation sequencing technology on genetics. Cell. 2008;24:133–141. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

