Molecular roles and function of circular RNAs in eukaryotic cells
- PMID: 29116363
- PMCID: PMC5814467
- DOI: 10.1007/s00018-017-2688-5
Molecular roles and function of circular RNAs in eukaryotic cells
Abstract
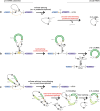
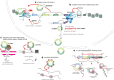
Protein-coding and noncoding genes in eukaryotes are typically expressed as linear messenger RNAs, with exons arranged colinearly to their genomic order. Recent advances in sequencing and in mapping RNA reads to reference genomes have revealed that thousands of genes express also covalently closed circular RNAs. Many of these circRNAs are stable and contain exons, but are not translated into proteins. Here, we review the emerging understanding that both, circRNAs produced by co- and posttranscriptional head-to-tail "backsplicing" of a downstream splice donor to a more upstream splice acceptor, as well as circRNAs generated from intronic lariats during colinear splicing, may exhibit physiologically relevant regulatory functions in eukaryotes. We describe how circRNAs impact gene expression of their host gene locus by affecting transcriptional initiation and elongation or splicing, and how they partake in controlling the function of other molecules, for example by interacting with microRNAs and proteins. We conclude with an outlook how circRNA dysregulation affects disease, and how the stability of circRNAs might be exploited in biomedical applications.
Keywords: Alu elements; Cancer; Cardiovascular disease; Chromatin; Exosomes; RNA polymerase II; Spliceosome; microRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

