Biological and phylogenetic characteristics of West African lineages of West Nile virus
- PMID: 29117195
- PMCID: PMC5695850
- DOI: 10.1371/journal.pntd.0006078
Biological and phylogenetic characteristics of West African lineages of West Nile virus
Abstract
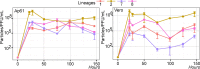
The West Nile virus (WNV), isolated in 1937, is an arbovirus (arthropod-borne virus) that infects thousands of people each year. Despite its burden on global health, little is known about the virus' biological and evolutionary dynamics. As several lineages are endemic in West Africa, we obtained the complete polyprotein sequence from three isolates from the early 1990s, each representing a different lineage. We then investigated differences in growth behavior and pathogenicity for four distinct West African lineages in arthropod (Ap61) and primate (Vero) cell lines, and in mice. We found that genetic differences, as well as viral-host interactions, could play a role in the biological properties in different WNV isolates in vitro, such as: (i) genome replication, (ii) protein translation, (iii) particle release, and (iv) virulence. Our findings demonstrate the endemic diversity of West African WNV strains and support future investigations into (i) the nature of WNV emergence, (ii) neurological tropism, and (iii) host adaptation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72: 73–83. Available: http://jvi.asm.org/content/72/1/73.short - PMC - PubMed
-
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44: 649–88. doi: 10.1146/annurev.mi.44.100190.003245 - DOI - PubMed
-
- Gould EA, Zanotto PM de A, Holmes EC. The genetic evolution of flaviviruses. 1997.
-
- Fall G, Diallo M, Loucoubar C, Faye O, Sall AA. Vector competence of Culex neavei and Culex quinquefasciatus (Diptera: Culicidae) from Senegal for lineages 1, 2, Koutango and a putative new lineage of West Nile virus. Am J Trop Med Hyg. 2014;90: 747–754. doi: 10.4269/ajtmh.13-0405 - DOI - PMC - PubMed
-
- Murgue B, Zeller H, Deubel V. The Ecology and Epidemiology of West Nile Virus in Africa, Europe and Asia In: Mackenzie JS, Barrett ADT, Deubel V, editors. Berlin, Heidelberg: Springer Berlin Heidelberg; 2002. pp. 195–221. doi: 10.1007/978-3-642-59403-8_10 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

