Genomic comparison between Staphylococcus aureus GN strains clinically isolated from a familial infection case: IS1272 transposition through a novel inverted repeat-replacing mechanism
- PMID: 29117225
- PMCID: PMC5678879
- DOI: 10.1371/journal.pone.0187288
Genomic comparison between Staphylococcus aureus GN strains clinically isolated from a familial infection case: IS1272 transposition through a novel inverted repeat-replacing mechanism
Abstract
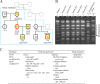
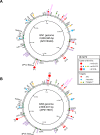
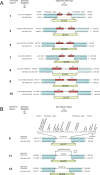
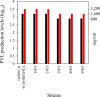
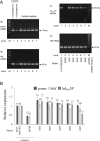
A bacterial insertion sequence (IS) is a mobile DNA sequence carrying only the transposase gene (tnp) that acts as a mutator to disrupt genes, alter gene expressions, and cause genomic rearrangements. "Canonical" ISs have historically been characterized by their terminal inverted repeats (IRs), which may form a stem-loop structure, and duplications of a short (non-IR) target sequence at both ends, called target site duplications (TSDs). The IS distributions and virulence potentials of Staphylococcus aureus genomes in familial infection cases are unclear. Here, we determined the complete circular genome sequences of familial strains from a Panton-Valentine leukocidin (PVL)-positive ST50/agr4 S. aureus (GN) infection of a 4-year old boy with skin abscesses. The genomes of the patient strain (GN1) and parent strain (GN3) were rich for "canonical" IS1272 with terminal IRs, both having 13 commonly-existing copies (ce-IS1272). Moreover, GN1 had a newly-inserted IS1272 (ni-IS1272) on the PVL-converting prophage, while GN3 had two copies of ni-IS1272 within the DNA helicase gene and near rot. The GN3 genome also had a small deletion. The targets of ni-IS1272 transposition were IR structures, in contrast with previous "canonical" ISs. There were no TSDs. Based on a database search, the targets for ce-IS1272 were IRs or "non-IRs". IS1272 included a larger structure with tandem duplications of the left (IRL) side sequence; tnp included minor cases of a long fusion form and truncated form. One ce-IS1272 was associated with the segments responsible for immune evasion and drug resistance. Regarding virulence, GN1 expressed cytolytic peptides (phenol-soluble modulin α and δ-hemolysin) and PVL more strongly than some other familial strains. These results suggest that IS1272 transposes through an IR-replacing mechanism, with an irreversible process unlike that of "canonical" transpositions, resulting in genomic variations, and that, among the familial strains, the patient strain has strong virulence potential based on community-associated virulence factors.
Conflict of interest statement
Figures









References
-
- Tacconelli E, Tumbarello M, Cauda R. Staphylococcus aureus infections. N Engl J Med. 1998; 339,2026–2027. - PubMed
-
- Tenover FC, Gorwitz RJ. The epidemiology of Staphylococcus infections In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-positive pathogens, 2nd Ed. Washington, DC: ASM Press; 2006. pp. 526–534.
-
- Powers ME, Bubeck Wardenburg J. Igniting the fire: Staphylococcus aureus virulence factors in the pathogenesis of sepsis. PLoS Pathog. 2014; 10:e1003871 doi: 10.1371/journal.ppat.1003871 - DOI - PMC - PubMed
-
- Kobayashi SD, Malachowa N, DeLeo FR. Pathogenesis of Staphylococcus aureus abscesses. Am J Pathol. 2015; 185:1518–1527. doi: 10.1016/j.ajpath.2014.11.030 - DOI - PMC - PubMed
-
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015; 28,603–661. doi: 10.1128/CMR.00134-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

