Structure-based discovery of cyclin-dependent protein kinase inhibitors
- PMID: 29118092
- PMCID: PMC6248306
- DOI: 10.1042/EBC20170040
Structure-based discovery of cyclin-dependent protein kinase inhibitors
Abstract
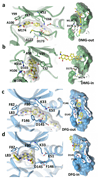
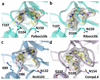
The cell fate-determining roles played by members of the cyclin-dependent protein kinase (CDK) family explain why their dysregulation can promote proliferative diseases, and identify them as potential targets for drug discovery in oncology and beyond. After many years of research, the first efficacious CDK inhibitors have now been registered for clinical use in a defined segment of breast cancer. Research is underway to identify inhibitors with appropriate CDK-inhibitory profiles to recapitulate this success in other disease settings. Here, we review the structural data that illustrate the interactions and properties that confer upon inhibitors affinity and/or selectivity toward different CDK family members. We conclude that where CDK inhibitors display selectivity, that selectivity derives from exploiting active site sequence peculiarities and/or from the capacity of the target CDK(s) to access conformations compatible with optimizing inhibitor-target interactions.
Keywords: CDK; Structure-based drug design; cell cycle; kinase.
© 2017 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


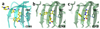

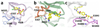

References
-
- Morgan DO. The Cell Cycle Principles of Control (Primers in Biology) New Science Press Ltd; 2007.
-
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

