SIRT1-dependent modulation of methylation and acetylation of histone H3 on lysine 9 (H3K9) in the zygotic pronuclei improves porcine embryo development
- PMID: 29118980
- PMCID: PMC5664433
- DOI: 10.1186/s40104-017-0214-0
SIRT1-dependent modulation of methylation and acetylation of histone H3 on lysine 9 (H3K9) in the zygotic pronuclei improves porcine embryo development
Abstract
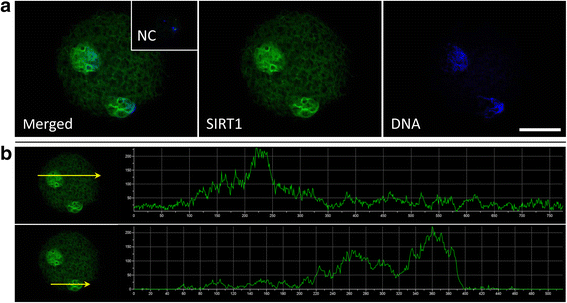
Background: The histone code is an established epigenetic regulator of early embryonic development in mammals. The lysine residue K9 of histone H3 (H3K9) is a prime target of SIRT1, a member of NAD+-dependent histone deacetylase family of enzymes targeting both histone and non-histone substrates. At present, little is known about SIRT1-modulation of H3K9 in zygotic pronuclei and its association with the success of preimplantation embryo development. Therefore, we evaluated the effect of SIRT1 activity on H3K9 methylation and acetylation in porcine zygotes and the significance of H3K9 modifications for early embryonic development.
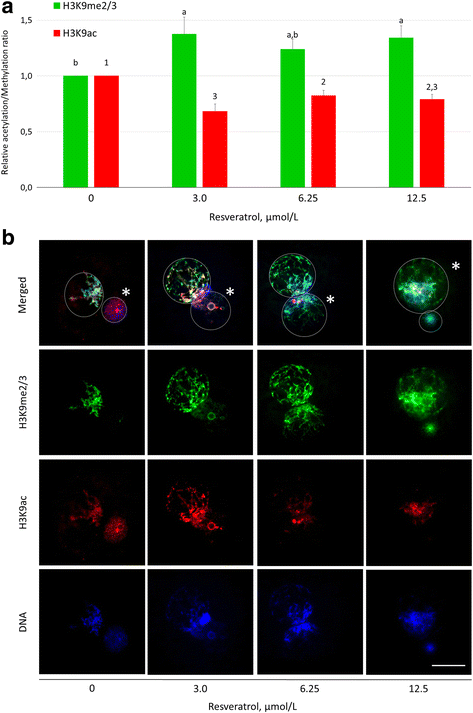
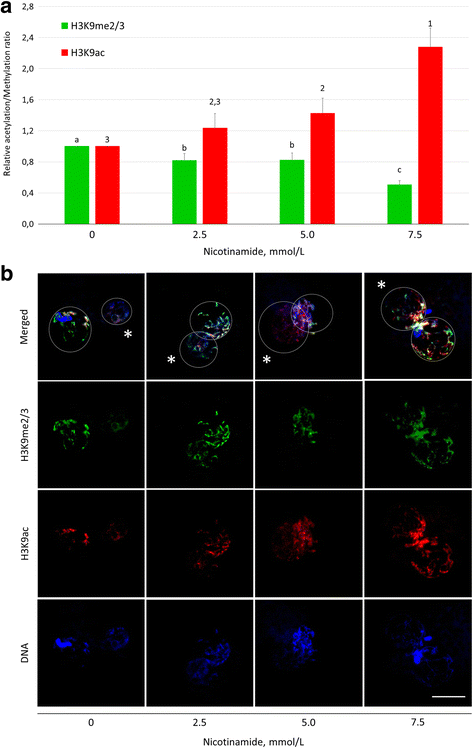
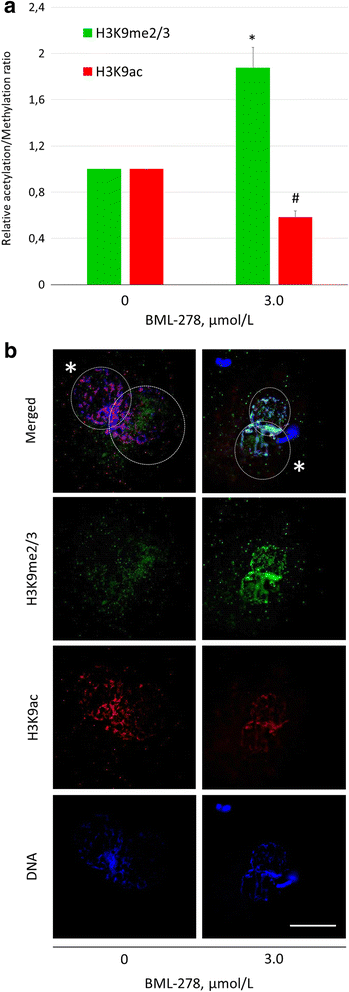
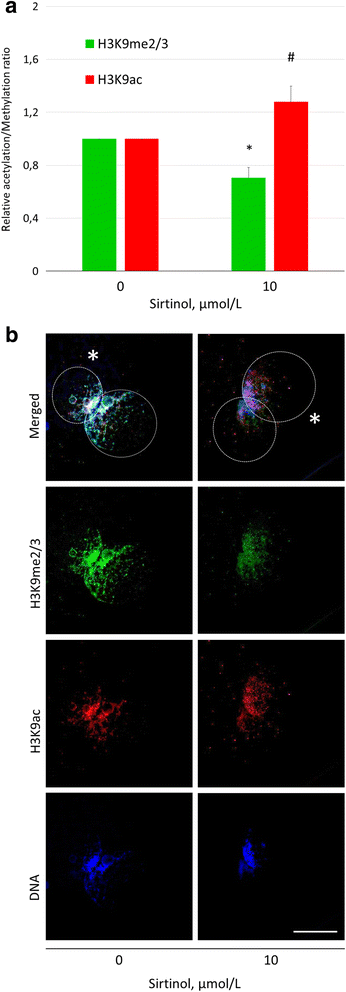
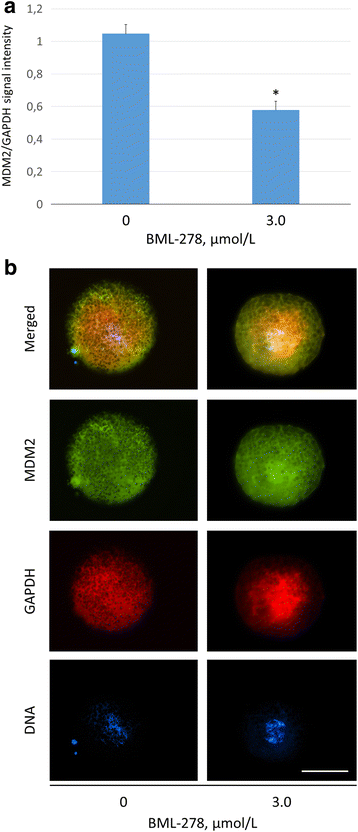
Results: Our results show that SIRT1 activators resveratrol and BML-278 increased H3K9 methylation and suppressed H3K9 acetylation in both the paternal and maternal pronucleus. Inversely, SIRT1 inhibitors nicotinamide and sirtinol suppressed methylation and increased acetylation of pronuclear H3K9. Evaluation of early embryonic development confirmed positive effect of selective SIRT1 activation on blastocyst formation rate (5.2 ± 2.9% versus 32.9 ± 8.1% in vehicle control and BML-278 group, respectively; P ≤ 0.05). Stimulation of SIRT1 activity coincided with fluorometric signal intensity of ooplasmic ubiquitin ligase MDM2, a known substrate of SIRT1 and known limiting factor of epigenome remodeling.
Conclusions: We conclude that SIRT1 modulates zygotic histone code, obviously through direct deacetylation and via non-histone targets resulting in increased H3K9me3. These changes in zygotes lead to more successful pre-implantation embryonic development and, indeed, the specific SIRT1 activation due to BML-278 is beneficial for in vitro embryo production and blastocyst achievement.
Keywords: Embryonic development; Epigenetics; H3K9 methylation; SIRT1; Sirtuin.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

