Bortezomib-induced miRNAs direct epigenetic silencing of locus genes and trigger apoptosis in leukemia
- PMID: 29120412
- PMCID: PMC5775404
- DOI: 10.1038/cddis.2017.520
Bortezomib-induced miRNAs direct epigenetic silencing of locus genes and trigger apoptosis in leukemia
Abstract
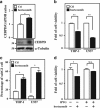
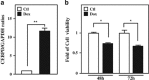
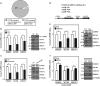
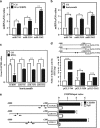
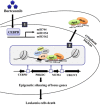
MicroRNAs (miRNAs) have been suggested to repress transcription via binding the 3'-untranslated regions of mRNAs. However, the involvement and details of miRNA-mediated epigenetic regulation, particularly in targeting genomic DNA and mediating epigenetic regulation, remain largely uninvestigated. In the present study, transcription factor CCAAT/enhancer binding protein delta (CEBPD) was responsive to the anticancer drug bortezomib, a clinical and highly selective drug for leukemia treatment, and contributed to bortezomib-induced cell death. Interestingly, following the identification of CEBPD-induced miRNAs, we found that miR-744, miR-3154 and miR-3162 could target CpG islands in the 5'-flanking region of the CEBPD gene. We previously demonstrated that the Yin Yang 1 (YY1)/polycomb group (PcG) protein/DNA methyltransferase (DNMT) complex is important for CCAAT/enhancer binding protein delta (CEBPD) gene inactivation; we further found that Argonaute 2 (Ago2) interacts with YY1 and binds to the CEBPD promoter. The miRNA/Ago2/YY1/PcG group protein/DNMT complex linked the inactivation of CEBPD and genes adjacent to its 5'-flanking region, including protein kinase DNA-activated catalytic polypeptide (PRKDC), minichromosome maintenance-deficient 4 (MCM4) and ubiquitin-conjugating enzyme E2 variant 2 (UBE2V2), upon bortezomib treatment. Moreover, we revealed that miRNA binding is necessary for YY1/PcG group protein/DNMT complex-mediated epigenetic gene silencing and is associated with bortezomib-induced methylation on genomic DNA. The present study successfully characterized the interactions of the miRNA/Ago2/YY1/PcG group protein/DNMT complex and provided new insights for miRNA-mediated epigenetic regulation in bortezomib-induced leukemic cell arrest and cell death.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex.J Biol Chem. 2008 Nov 7;283(45):30919-32. doi: 10.1074/jbc.M804029200. Epub 2008 Aug 27. J Biol Chem. 2008. PMID: 18753137 Free PMC article.
-
Yin Yang 1-mediated epigenetic silencing of tumour-suppressive microRNAs activates nuclear factor-κB in hepatocellular carcinoma.J Pathol. 2016 Apr;238(5):651-64. doi: 10.1002/path.4688. J Pathol. 2016. PMID: 26800240
-
MiR-193b Mediates CEBPD-Induced Cisplatin Sensitization Through Targeting ETS1 and Cyclin D1 in Human Urothelial Carcinoma Cells.J Cell Biochem. 2017 Jun;118(6):1563-1573. doi: 10.1002/jcb.25818. Epub 2016 Dec 20. J Cell Biochem. 2017. PMID: 27918099
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
Cited by
-
Bortezomib-Induced Epigenetic Alterations in Nerve Cells: Focus on the Mechanisms Contributing to the Peripheral Neuropathy Development.Int J Mol Sci. 2022 Feb 23;23(5):2431. doi: 10.3390/ijms23052431. Int J Mol Sci. 2022. PMID: 35269574 Free PMC article.
-
Molecular Mechanisms of Bortezomib Action: Novel Evidence for the miRNA-mRNA Interaction Involvement.Int J Mol Sci. 2020 Jan 5;21(1):350. doi: 10.3390/ijms21010350. Int J Mol Sci. 2020. PMID: 31948068 Free PMC article.
-
Hypomethylating Chemotherapeutic Agents as Therapy for Myelodysplastic Syndromes and Prevention of Acute Myeloid Leukemia.Pharmaceuticals (Basel). 2021 Jul 4;14(7):641. doi: 10.3390/ph14070641. Pharmaceuticals (Basel). 2021. PMID: 34358067 Free PMC article. Review.
-
Epigenetic Alterations as Vital Aspects of Bortezomib Molecular Action.Cancers (Basel). 2023 Dec 23;16(1):84. doi: 10.3390/cancers16010084. Cancers (Basel). 2023. PMID: 38201512 Free PMC article. Review.
-
In situ Electrochemical Evaluation of the Interaction of dsDNA with the Proteasome Inhibitor Anticancer Drug Bortezomib.Molecules. 2023 Apr 6;28(7):3277. doi: 10.3390/molecules28073277. Molecules. 2023. PMID: 37050039 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

