Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion
- PMID: 29121062
- PMCID: PMC5679612
- DOI: 10.1371/journal.pone.0176181
Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion
Abstract
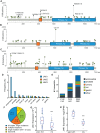
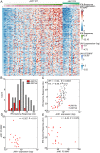
Immune evasion is a well-recognized hallmark of cancer and recent studies with immunotherapy agents have suggested that tumors with increased numbers of neoantigens elicit greater immune responses. We hypothesized that the immune system presents a common selective pressure on high mutation burden tumors and therefore immune evasion mutations would be enriched in high mutation burden tumors. The JAK family of kinases is required for the signaling of a host of immune modulators in tumor, stromal, and immune cells. Therefore, we analyzed alterations in this family for the hypothesized signature of an immune evasion mutation. Here, we searched a database of 61,704 unique solid tumors for alterations in the JAK family kinases (JAK1/2/3, TYK2). We used The Cancer Genome Atlas and Cancer Cell Line Encyclopedia data to confirm and extend our findings by analyzing gene expression patterns. Recurrent frameshift mutations in JAK1 were associated with high mutation burden and microsatellite instability. These mutations occurred in multiple tumor types including endometrial, colorectal, stomach, and prostate carcinomas. Analyzing gene expression signatures in endometrial and stomach adenocarcinomas revealed that tumors with a JAK1 frameshift exhibited reduced expression of interferon response signatures and multiple anti-tumor immune signatures. Importantly, endometrial cancer cell lines exhibited similar gene expression changes that were expected to be tumor cell intrinsic (e.g. interferon response) but not those expected to be tumor cell extrinsic (e.g. NK cells). From these data, we derive two primary conclusions: 1) JAK1 frameshifts are loss of function alterations that represent a potential pan-cancer adaptation to immune responses against tumors with microsatellite instability; 2) The mechanism by which JAK1 loss of function contributes to tumor immune evasion is likely associated with loss of the JAK1-mediated interferon response.
Conflict of interest statement
Figures




References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013 - DOI - PubMed
-
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–8. doi: 10.1038/ni1102-991 - DOI - PubMed
-
- Clemente CG, Mihm MC Jr., Bufalino R, Zurrida S, Collini P, Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77(7):1303–10. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1303::AID-CNCR12>3.0.CO;2-5 - DOI - PubMed
-
- Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–13. doi: 10.1056/NEJMoa020177 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

