Nature is the best source of anticancer drugs: Indexing natural products for their anticancer bioactivity
- PMID: 29121120
- PMCID: PMC5679595
- DOI: 10.1371/journal.pone.0187925
Nature is the best source of anticancer drugs: Indexing natural products for their anticancer bioactivity
Abstract
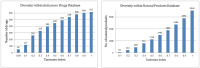
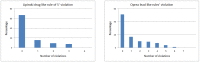
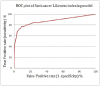
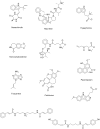
Cancer is considered one of the primary diseases that cause morbidity and mortality in millions of people worldwide and due to its prevalence, there is undoubtedly an unmet need to discover novel anticancer drugs. However, the traditional process of drug discovery and development is lengthy and expensive, so the application of in silico techniques and optimization algorithms in drug discovery projects can provide a solution, saving time and costs. A set of 617 approved anticancer drugs, constituting the active domain, and a set of 2,892 natural products, constituting the inactive domain, were employed to build predictive models and to index natural products for their anticancer bioactivity. Using the iterative stochastic elimination optimization technique, we obtained a highly discriminative and robust model, with an area under the curve of 0.95. Twelve natural products that scored highly as potential anticancer drug candidates are disclosed. Searching the scientific literature revealed that few of those molecules (Neoechinulin, Colchicine, and Piperolactam) have already been experimentally screened for their anticancer activity and found active. The other phytochemicals await evaluation for their anticancerous activity in wet lab.
Conflict of interest statement
Figures
References
-
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128. doi: 10.1016/S0140-6736(12)61728-0 . - DOI - PMC - PubMed
-
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 . - DOI - PubMed
-
- Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330(6009):1340–4. doi: 10.1126/science.1193494 . - DOI - PubMed
-
- Ngo DC, Ververis K, Tortorella SM, Karagiannis TC. Introduction to the molecular basis of cancer metabolism and the Warburg effect. Mol Biol Rep. 2015;42(4):819–23. doi: 10.1007/s11033-015-3857-y . - DOI - PubMed
-
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33. doi: 10.1126/science.1160809 . - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

