PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1
- PMID: 29121337
- PMCID: PMC5728402
- DOI: 10.1093/nar/gkx1047
PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1
Abstract
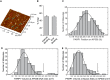
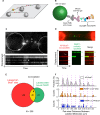
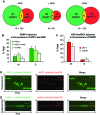
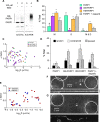
PARP1-dependent poly-ADP-ribosylation (PARylation) participates in the repair of many forms of DNA damage. Here, we used atomic force microscopy (AFM) and single molecule fluorescence microscopy to examine the interactions of PARP1 with common DNA repair intermediates. AFM volume analysis indicates that PARP1 binds to DNA at nicks, abasic (AP) sites, and ends as a monomer. Single molecule DNA tightrope assays were used to follow the real-time dynamic behavior of PARP1 in the absence and presence of AP endonuclease (APE1) on AP DNA damage arrays. These experiments revealed that PARP1 conducted damage search mostly through 3D diffusion. Co-localization of APE1 with PARP1 on DNA was found capable of inducing 1D diffusion of otherwise nonmotile PARP1, while excess APE1 also facilitated the dissociation of DNA-bound PARP1. Moreover, auto-PARylation of PARP1 allowed the protein to switch its damage search strategy by causing a 3-fold increase in linear diffusion. Finally, we demonstrated that PARP inhibitor olaparib did not significantly alter the rate of PARP1 dissociation from DNA, but instead resulted in more motility of DNA-bound PARP1 molecules.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Ame J.C., Spenlehauer C., de Murcia G.. The PARP superfamily. Bioessays. 2004; 26:882–893. - PubMed
-
- Benjamin R.C., Gill D.M.. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J. Biol. Chem. 1980; 255:10502–10508. - PubMed
-
- Dantzer F., Ame J.C., Schreiber V., Nakamura J., Menissier-de Murcia J., de Murcia G.. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006; 409:493–510. - PubMed
-
- Pion E., Ullmann G.M., Ame J.C., Gerard D., de Murcia G., Bombarda E.. DNA-induced dimerization of poly(ADP-ribose) polymerase-1 triggers its activation. Biochemistry. 2005; 44:14670–14681. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

