Secondary structure forming sequences drive SD-MMEJ repair of DNA double-strand breaks
- PMID: 29121353
- PMCID: PMC5728401
- DOI: 10.1093/nar/gkx1056
Secondary structure forming sequences drive SD-MMEJ repair of DNA double-strand breaks
Abstract
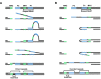
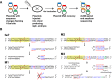
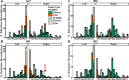
Alternative end-joining (alt-EJ) repair of DNA double-strand breaks is associated with deletions, chromosome translocations, and genome instability. Alt-EJ frequently uses annealing of microhomologous sequences to tether broken ends. When accessible pre-existing microhomologies do not exist, we have postulated that new microhomologies can be created via limited DNA synthesis at secondary-structure forming sequences. This model, called synthesis-dependent microhomology-mediated end joining (SD-MMEJ), predicts that differences between DNA sequences near double-strand breaks should alter repair outcomes in predictable ways. To test this hypothesis, we injected plasmids with sequence variations flanking an I-SceI endonuclease recognition site into I-SceI expressing Drosophila embryos and used Illumina amplicon sequencing to compare repair junctions. As predicted by the model, we found that small changes in sequences near the I-SceI site had major impacts on the spectrum of repair junctions. Bioinformatic analyses suggest that these repair differences arise from transiently forming loops and hairpins within 30 nucleotides of the break. We also obtained evidence for 'trans SD-MMEJ,' involving at least two consecutive rounds of microhomology annealing and synthesis across the break site. These results highlight the importance of sequence context for alt-EJ repair and have important implications for genome editing and genome evolution.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
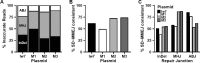




Similar articles
-
Characterization of sequence contexts that favor alternative end joining at Cas9-induced double-strand breaks.Nucleic Acids Res. 2022 Jul 22;50(13):7465-7478. doi: 10.1093/nar/gkac575. Nucleic Acids Res. 2022. PMID: 35819195 Free PMC article.
-
Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions.Nucleic Acids Res. 2010 Sep;38(17):5706-17. doi: 10.1093/nar/gkq379. Epub 2010 May 11. Nucleic Acids Res. 2010. PMID: 20460465 Free PMC article.
-
Recovery of Alternative End-Joining Repair Products From Drosophila Embryos.Methods Enzymol. 2018;601:91-110. doi: 10.1016/bs.mie.2017.11.027. Epub 2018 Feb 3. Methods Enzymol. 2018. PMID: 29523244
-
Risky business: Microhomology-mediated end joining.Mutat Res. 2016 Jun;788:17-24. doi: 10.1016/j.mrfmmm.2015.12.005. Epub 2016 Jan 2. Mutat Res. 2016. PMID: 26790771 Free PMC article. Review.
-
Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway?Trends Biochem Sci. 2015 Nov;40(11):701-714. doi: 10.1016/j.tibs.2015.08.006. Epub 2015 Oct 1. Trends Biochem Sci. 2015. PMID: 26439531 Free PMC article. Review.
Cited by
-
Unravelling roles of error-prone DNA polymerases in shaping cancer genomes.Oncogene. 2021 Dec;40(48):6549-6565. doi: 10.1038/s41388-021-02032-9. Epub 2021 Oct 18. Oncogene. 2021. PMID: 34663880 Free PMC article. Review.
-
Mechanistic basis for microhomology identification and genome scarring by polymerase theta.Proc Natl Acad Sci U S A. 2020 Apr 14;117(15):8476-8485. doi: 10.1073/pnas.1921791117. Epub 2020 Mar 31. Proc Natl Acad Sci U S A. 2020. PMID: 32234782 Free PMC article.
-
A Dual sgRNA Approach for Functional Genomics in Arabidopsis thaliana.G3 (Bethesda). 2018 Jul 31;8(8):2603-2615. doi: 10.1534/g3.118.200046. G3 (Bethesda). 2018. PMID: 29884615 Free PMC article.
-
Parp3 promotes long-range end joining in murine cells.Proc Natl Acad Sci U S A. 2018 Oct 2;115(40):10076-10081. doi: 10.1073/pnas.1801591115. Epub 2018 Sep 13. Proc Natl Acad Sci U S A. 2018. PMID: 30213852 Free PMC article.
-
Characterization of sequence contexts that favor alternative end joining at Cas9-induced double-strand breaks.Nucleic Acids Res. 2022 Jul 22;50(13):7465-7478. doi: 10.1093/nar/gkac575. Nucleic Acids Res. 2022. PMID: 35819195 Free PMC article.
References
-
- Lieber M.R., Ma Y., Pannicke U., Schwarz K.. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 2003; 4:712–720. - PubMed
-
- Lieber M.R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008; 283:1–5. - PubMed
-
- Lieber M.R., Lu H., Gu J., Schwarz K.. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008; 18:125–133. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

