Investigating the hepatitis B virus life cycle using engineered reporter hepatitis B viruses
- PMID: 29121422
- PMCID: PMC5765299
- DOI: 10.1111/cas.13440
Investigating the hepatitis B virus life cycle using engineered reporter hepatitis B viruses
Abstract
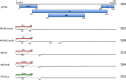
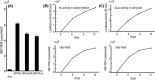
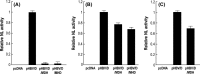
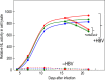
Chronic infection with hepatitis B virus (HBV) increases the risk of developing fibrosis, cirrhosis or hepatocellular carcinoma. Current therapies are limited to type-I interferons and/or nucleos(t)ide analogues; however, these are only partially effective. The development of novel anti-HBV agents for new treatment strategies has been hampered by the lack of a suitable system that allows the in vitro replication of HBV. Studies of virus infection/replication at the molecular level using wild-type HBV are labor-intensive and time-consuming. To overcome these problems, we previously constructed a recombinant reporter HBV bearing the NanoLuc gene and showed its usefulness in identifying factors that affect HBV proliferation. Because this system mimics the early stage of the HBV life cycle faithfully, we conducted a quantitative analysis of HBV infectivity to several human hepatocyte cell lines as well as the effect of dimethyl sulfoxide and HBV protein X on the early stage of HBV proliferation using this system. Furthermore, we developed a system to produce a reporter HBV expressing a pol gene. These reporter HBV may provide an opportunity to enhance our understanding of the HBV life cycle and aid strategies for the development of new anti-HBV agents.
Keywords: anti- hepatitis B virus agents; hepatitis B virus; replication; reporter gene; reporter hepatitis B virus.
© 2017 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures




Similar articles
-
Construction of a replication-competent hepatitis B virus vector carrying secreted luciferase transgene and establishment of new hepatitis B virus replication and expression cell lines.World J Gastroenterol. 2019 Oct 21;25(39):5961-5972. doi: 10.3748/wjg.v25.i39.5961. World J Gastroenterol. 2019. PMID: 31660033 Free PMC article.
-
Robust Human and Murine Hepatocyte Culture Models of Hepatitis B Virus Infection and Replication.J Virol. 2018 Nov 12;92(23):e01255-18. doi: 10.1128/JVI.01255-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30232184 Free PMC article.
-
Development of a Hepatitis B Virus Reporter System to Monitor the Early Stages of the Replication Cycle.J Vis Exp. 2017 Feb 1;(120):54849. doi: 10.3791/54849. J Vis Exp. 2017. PMID: 28190076 Free PMC article.
-
HBx protein as a therapeutic target for functional cure of hepatitis B virus infection.Virology. 2025 Mar;604:110438. doi: 10.1016/j.virol.2025.110438. Epub 2025 Jan 31. Virology. 2025. PMID: 39908774 Review.
-
Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma.Microb Pathog. 2019 Mar;128:184-194. doi: 10.1016/j.micpath.2019.01.004. Epub 2019 Jan 3. Microb Pathog. 2019. PMID: 30611768 Review.
Cited by
-
Mapping the Interactions of HBV cccDNA with Host Factors.Int J Mol Sci. 2019 Sep 1;20(17):4276. doi: 10.3390/ijms20174276. Int J Mol Sci. 2019. PMID: 31480501 Free PMC article. Review.
-
Utility of Three-Dimensional Cultures of Primary Human Hepatocytes (Spheroids) as Pharmacokinetic Models.Biomedicines. 2020 Sep 23;8(10):374. doi: 10.3390/biomedicines8100374. Biomedicines. 2020. PMID: 32977664 Free PMC article. Review.
-
Cell and Animal Models for Studying Hepatitis B Virus Infection and Drug Development.Gastroenterology. 2019 Jan;156(2):338-354. doi: 10.1053/j.gastro.2018.06.093. Epub 2018 Sep 19. Gastroenterology. 2019. PMID: 30243619 Free PMC article. Review.
-
MicroRNA-3145 as a potential therapeutic target for hepatitis B virus: inhibition of viral replication via downregulation of HBS and HBX.Front Microbiol. 2025 Jan 6;15:1499216. doi: 10.3389/fmicb.2024.1499216. eCollection 2024. Front Microbiol. 2025. PMID: 39834379 Free PMC article.
-
Neutralization of hepatitis B virus with vaccine-escape mutations by hepatitis B vaccine with large-HBs antigen.Nat Commun. 2022 Sep 5;13(1):5207. doi: 10.1038/s41467-022-32910-z. Nat Commun. 2022. PMID: 36064848 Free PMC article.
References
-
- WHO . Hepatitis B. WHO Hepatitis B Fact sheet no 204, 2008.
-
- Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235‐249. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

