Coculture of Marine Invertebrate-Associated Bacteria and Interdisciplinary Technologies Enable Biosynthesis and Discovery of a New Antibiotic, Keyicin
- PMID: 29121465
- PMCID: PMC5973552
- DOI: 10.1021/acschembio.7b00688
Coculture of Marine Invertebrate-Associated Bacteria and Interdisciplinary Technologies Enable Biosynthesis and Discovery of a New Antibiotic, Keyicin
Abstract
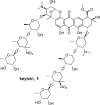
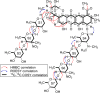
Advances in genomics and metabolomics have made clear in recent years that microbial biosynthetic capacities on Earth far exceed previous expectations. This is attributable, in part, to the realization that most microbial natural product (NP) producers harbor biosynthetic machineries not readily amenable to classical laboratory fermentation conditions. Such "cryptic" or dormant biosynthetic gene clusters (BGCs) encode for a vast assortment of potentially new antibiotics and, as such, have become extremely attractive targets for activation under controlled laboratory conditions. We report here that coculturing of a Rhodococcus sp. and a Micromonospora sp. affords keyicin, a new and otherwise unattainable bis-nitroglycosylated anthracycline whose mechanism of action (MOA) appears to deviate from those of other anthracyclines. The structure of keyicin was elucidated using high resolution MS and NMR technologies, as well as detailed molecular modeling studies. Sequencing of the keyicin BGC (within the Micromonospora genome) enabled both structural and genomic comparisons to other anthracycline-producing systems informing efforts to characterize keyicin. The new NP was found to be selectively active against Gram-positive bacteria including both Rhodococcus sp. and Mycobacterium sp. E. coli-based chemical genomics studies revealed that keyicin's MOA, in contrast to many other anthracyclines, does not invoke nucleic acid damage.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

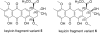
Similar articles
-
Omics Technologies to Understand Activation of a Biosynthetic Gene Cluster in Micromonospora sp. WMMB235: Deciphering Keyicin Biosynthesis.ACS Chem Biol. 2019 Jun 21;14(6):1260-1270. doi: 10.1021/acschembio.9b00223. Epub 2019 Jun 7. ACS Chem Biol. 2019. PMID: 31120241 Free PMC article.
-
A new anthracycline antibiotic micromonomycin from Micromonospora sp.J Antibiot (Tokyo). 2004 Sep;57(9):601-4. doi: 10.7164/antibiotics.57.601. J Antibiot (Tokyo). 2004. PMID: 15580962 No abstract available.
-
Characterization of the biosynthetic gene cluster for maklamicin, a spirotetronate-class antibiotic of the endophytic Micromonospora sp. NBRC 110955.Microbiol Res. 2015 Nov;180:30-9. doi: 10.1016/j.micres.2015.07.003. Epub 2015 Jul 22. Microbiol Res. 2015. PMID: 26505309
-
From chemical structure to environmental biosynthetic pathways: navigating marine invertebrate-bacteria associations.Trends Biotechnol. 2005 Sep;23(9):437-40. doi: 10.1016/j.tibtech.2005.07.002. Trends Biotechnol. 2005. PMID: 16038996 Review.
-
The regulation of the secondary metabolism of Streptomyces: new links and experimental advances.Nat Prod Rep. 2011 Jul;28(7):1311-33. doi: 10.1039/c1np00003a. Epub 2011 May 25. Nat Prod Rep. 2011. PMID: 21611665 Review.
Cited by
-
Pathway Engineering of Anthracyclines: Blazing Trails in Natural Product Glycodiversification.J Org Chem. 2020 Oct 2;85(19):12012-12023. doi: 10.1021/acs.joc.0c01863. Epub 2020 Sep 22. J Org Chem. 2020. PMID: 32938175 Free PMC article.
-
Bacteria Cultivated From Sponges and Bacteria Not Yet Cultivated From Sponges-A Review.Front Microbiol. 2021 Nov 10;12:737925. doi: 10.3389/fmicb.2021.737925. eCollection 2021. Front Microbiol. 2021. PMID: 34867854 Free PMC article. Review.
-
Discovery of novel secondary metabolites encoded in actinomycete genomes through coculture.J Ind Microbiol Biotechnol. 2021 Jun 4;48(3-4):kuaa001. doi: 10.1093/jimb/kuaa001. J Ind Microbiol Biotechnol. 2021. PMID: 33825906 Free PMC article. Review.
-
Elicitation for activation of the actinomycete genome's cryptic secondary metabolite gene clusters.RSC Adv. 2023 Feb 16;13(9):5778-5795. doi: 10.1039/d2ra08222e. eCollection 2023 Feb 14. RSC Adv. 2023. PMID: 36816076 Free PMC article. Review.
-
Response of Secondary Metabolism of Hypogean Actinobacterial Genera to Chemical and Biological Stimuli.Appl Environ Microbiol. 2018 Sep 17;84(19):e01125-18. doi: 10.1128/AEM.01125-18. Print 2018 Oct 1. Appl Environ Microbiol. 2018. PMID: 30030223 Free PMC article.
References
-
- Shih CJ, Chen PY, Liaw CC, Lai YM, Yang YL. Bringing microbial interactions to light using imaging mass spectrometry. Nat Prod Rep. 2014;31:739–755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008505/GM/NIGMS NIH HHS/United States
- R01 HG005084/HG/NHGRI NIH HHS/United States
- UL1 TR000427/TR/NCATS NIH HHS/United States
- T32 GM008347/GM/NIGMS NIH HHS/United States
- U19 AI109673/AI/NIAID NIH HHS/United States
- S10 RR029531/RR/NCRR NIH HHS/United States
- R01 GM104192/GM/NIGMS NIH HHS/United States
- S10 RR002781/RR/NCRR NIH HHS/United States
- P41 GM103399/GM/NIGMS NIH HHS/United States
- R01 DK071801/DK/NIDDK NIH HHS/United States
- S10 RR008438/RR/NCRR NIH HHS/United States
- R01 GM104975/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

