The combinatorial control of alternative splicing in C. elegans
- PMID: 29121637
- PMCID: PMC5697891
- DOI: 10.1371/journal.pgen.1007033
The combinatorial control of alternative splicing in C. elegans
Abstract
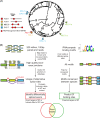
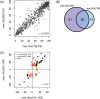
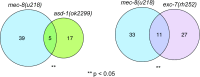
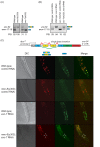
Normal development requires the right splice variants to be made in the right tissues at the right time. The core splicing machinery is engaged in all splicing events, but which precise splice variant is made requires the choice between alternative splice sites-for this to occur, a set of splicing factors (SFs) must recognize and bind to short RNA motifs in the pre-mRNA. In C. elegans, there is known to be extensive variation in splicing patterns across development, but little is known about the targets of each SF or how multiple SFs combine to regulate splicing. Here we combine RNA-seq with in vitro binding assays to study how 4 different C. elegans SFs, ASD-1, FOX-1, MEC-8, and EXC-7, regulate splicing. The 4 SFs chosen all have well-characterised biology and well-studied loss-of-function genetic alleles, and all contain RRM domains. Intriguingly, while the SFs we examined have varied roles in C. elegans development, they show an unexpectedly high overlap in their targets. We also find that binding sites for these SFs occur on the same pre-mRNAs more frequently than expected suggesting extensive combinatorial control of splicing. We confirm that regulation of splicing by multiple SFs is often combinatorial and show that this is functionally significant. We also find that SFs appear to combine to affect splicing in two modes-they either bind in close proximity within the same intron or they appear to bind to separate regions of the intron in a conserved order. Finally, we find that the genes whose splicing are regulated by multiple SFs are highly enriched for genes involved in the cytoskeleton and in ion channels that are key for neurotransmission. Together, this shows that specific classes of genes have complex combinatorial regulation of splicing and that this combinatorial regulation is critical for normal development to occur.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Muscle-specific splicing factors ASD-2 and SUP-12 cooperatively switch alternative pre-mRNA processing patterns of the ADF/cofilin gene in Caenorhabditis elegans.PLoS Genet. 2012;8(10):e1002991. doi: 10.1371/journal.pgen.1002991. Epub 2012 Oct 11. PLoS Genet. 2012. PMID: 23071450 Free PMC article.
-
The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans.J Cell Biol. 2004 Nov 22;167(4):639-47. doi: 10.1083/jcb.200407085. Epub 2004 Nov 15. J Cell Biol. 2004. PMID: 15545320 Free PMC article.
-
The multiplicity of alternative splicing decisions in Caenorhabditis elegans is linked to specific intronic regulatory motifs and minisatellites.BMC Genomics. 2014 May 14;15(1):364. doi: 10.1186/1471-2164-15-364. BMC Genomics. 2014. PMID: 24884695 Free PMC article.
-
Alternative splicing in C. elegans.WormBook. 2005 Sep 26:1-13. doi: 10.1895/wormbook.1.31.1. WormBook. 2005. PMID: 18050427 Free PMC article. Review.
-
Pre-mRNA splicing and its regulation in Caenorhabditis elegans.WormBook. 2012 Mar 21:1-21. doi: 10.1895/wormbook.1.31.2. WormBook. 2012. PMID: 22467343 Free PMC article. Review.
Cited by
-
Identify differential genes and cell subclusters from time-series scRNA-seq data using scTITANS.Comput Struct Biotechnol J. 2021 Jul 26;19:4132-4141. doi: 10.1016/j.csbj.2021.07.016. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34527187 Free PMC article.
-
Detecting gene expression in Caenorhabditis elegans.Genetics. 2025 Jan 8;229(1):1-108. doi: 10.1093/genetics/iyae167. Genetics. 2025. PMID: 39693264 Free PMC article. Review.
-
Sensory neuron transcriptomes reveal complex neuron-specific function and regulation of mec-2/Stomatin splicing.Nucleic Acids Res. 2022 Mar 21;50(5):2401-2416. doi: 10.1093/nar/gkab1134. Nucleic Acids Res. 2022. PMID: 34875684 Free PMC article.
-
Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals.Genome Biol. 2021 Jan 14;22(1):35. doi: 10.1186/s13059-020-02258-y. Genome Biol. 2021. PMID: 33446251 Free PMC article.
-
Alternative splicing across the C. elegans nervous system.Nat Commun. 2025 May 16;16(1):4508. doi: 10.1038/s41467-025-58293-5. Nat Commun. 2025. PMID: 40379606 Free PMC article.
References
-
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12: 715–729. doi: 10.1038/nrg3052 - DOI - PMC - PubMed
-
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152: 1252–1269. doi: 10.1016/j.cell.2013.02.034 - DOI - PMC - PubMed
-
- Gracida X, Norris AD, Calarco JA. Regulation of Tissue-Specific Alternative Splicing: C. elegans as a Model System. Adv Exp Med Biol. 2016;907: 229–261. doi: 10.1007/978-3-319-29073-7_10 - DOI - PubMed
-
- Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11: 345–355. doi: 10.1038/nrg2776 - DOI - PubMed
-
- Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14: 153–165. doi: 10.1038/nrm3525 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

