The helicase DinG responds to stress due to DNA double strand breaks
- PMID: 29121674
- PMCID: PMC5679670
- DOI: 10.1371/journal.pone.0187900
The helicase DinG responds to stress due to DNA double strand breaks
Abstract
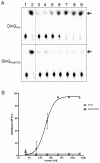
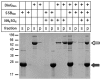
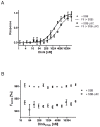
Neisseria meningitidis (Nm) is a Gram-negative nasopharyngeal commensal that can cause septicaemia and meningitis. The neisserial DNA damage-inducible protein DinG is a helicase related to the mammalian helicases XPD and FANCJ. These helicases belong to superfamily 2, are ATP dependent and exert 5' → 3' directionality. To better understand the role of DinG in neisserial genome maintenance, the Nm DinG (DinGNm) enzymatic activities were assessed in vitro and phenotypical characterization of a dinG null mutant (NmΔdinG) was performed. Like its homologues, DinGNm possesses 5' → 3' directionality and prefers DNA substrates containing a 5'-overhang. ATPase activity of DinGNm is strictly DNA-dependent and DNA unwinding activity requires nucleoside triphosphate and divalent metal cations. DinGNm directly binds SSBNm with a Kd of 313 nM. Genotoxic stress analysis demonstrated that NmΔdinG was more sensitive to double-strand DNA breaks (DSB) induced by mitomycin C (MMC) than the Nm wildtype, defining the role of neisserial DinG in DSB repair. Notably, when NmΔdinG cells grown under MMC stress assessed by quantitative mass spectrometry, 134 proteins were shown to be differentially abundant (DA) compared to unstressed NmΔdinG cells. Among the DNA replication, repair and recombination proteins affected, polymerase III subunits and recombinational repair proteins RuvA, RuvB, RecB and RecD were significantly down regulated while TopA and SSB were upregulated under stress condition. Most of the other DA proteins detected are involved in metabolic functions. The present study shows that the helicase DinG is probably involved in regulating metabolic pathways as well as in genome maintenance.
Conflict of interest statement
Figures







Similar articles
-
Characterization of the Neisseria meningitidis Helicase RecG.PLoS One. 2016 Oct 13;11(10):e0164588. doi: 10.1371/journal.pone.0164588. eCollection 2016. PLoS One. 2016. PMID: 27736945 Free PMC article.
-
Characterization of the DNA damage-inducible helicase DinG from Escherichia coli.J Biol Chem. 2003 Jul 25;278(30):28284-93. doi: 10.1074/jbc.M301188200. Epub 2003 May 14. J Biol Chem. 2003. PMID: 12748189
-
Impact of age-associated cyclopurine lesions on DNA repair helicases.PLoS One. 2014 Nov 19;9(11):e113293. doi: 10.1371/journal.pone.0113293. eCollection 2014. PLoS One. 2014. PMID: 25409515 Free PMC article.
-
The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes.DNA Repair (Amst). 2010 Mar 2;9(3):276-85. doi: 10.1016/j.dnarep.2009.12.016. Epub 2010 Jan 29. DNA Repair (Amst). 2010. PMID: 20116346 Review.
-
Structure, function and evolution of the bacterial DinG-like proteins.Comput Struct Biotechnol J. 2025 Mar 17;27:1124-1139. doi: 10.1016/j.csbj.2025.03.023. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40206346 Free PMC article. Review.
Cited by
-
Type IV CRISPR-Cas systems are highly diverse and involved in competition between plasmids.Nucleic Acids Res. 2020 Feb 28;48(4):2000-2012. doi: 10.1093/nar/gkz1197. Nucleic Acids Res. 2020. PMID: 31879772 Free PMC article.
-
Structural and functional investigation of DinG containing a 3'-5' exonuclease domain.mBio. 2025 Aug 13;16(8):e0088425. doi: 10.1128/mbio.00884-25. Epub 2025 Jun 30. mBio. 2025. PMID: 40586552 Free PMC article.
-
Serum proteases prevent bacterial biofilm formation: role of kallikrein and plasmin.Virulence. 2021 Dec;12(1):2902-2917. doi: 10.1080/21505594.2021.2003115. Virulence. 2021. PMID: 34903146 Free PMC article.
-
Alternative complexes formed by the Escherichia coli clamp loader accessory protein HolC (x) with replication protein HolD (ψ) and repair protein YoaA.DNA Repair (Amst). 2021 Apr;100:103006. doi: 10.1016/j.dnarep.2020.103006. Epub 2021 Feb 2. DNA Repair (Amst). 2021. PMID: 33582602 Free PMC article.
-
DNA repair is essential for Vibrio cholerae growth on Thiosulfate-Citrate-Bile Salts-Sucrose (TCBS) Medium.bioRxiv [Preprint]. 2025 Jan 11:2025.01.10.632459. doi: 10.1101/2025.01.10.632459. bioRxiv. 2025. Update in: J Bacteriol. 2025 Apr 17;207(4):e0000425. doi: 10.1128/jb.00004-25. PMID: 39829866 Free PMC article. Updated. Preprint.
References
-
- Nassif X. Microbiology. A furtive pathogen revealed. Science. 2000;287(5459):1767–8. Epub 2001/02/07 11:00. . - PubMed
-
- Davidsen T, Tonjum T. Meningococcal genome dynamics. Nat Rev Microbiol. 2006;4(1):11–22. Epub 2005/12/17 09:00. doi: 10.1038/nrmicro1324 . - DOI - PubMed
-
- Davidsen T, Tuven HK, Bjoras M, Rodland EA, Tonjum T. Genetic interactions of DNA repair pathways in the pathogen Neisseria meningitidis. J Bacteriol. 2007;189(15):5728–37. Epub 2007/05/22 09:00. doi: 10.1128/JB.00161-07 . - DOI - PMC - PubMed
-
- Raney KD, Byrd AK, Aarattuthodiyil S. Structure and Mechanisms of SF1 DNA Helicases. Adv Exp Med Biol. 2013;973(10):E1 Epub 2012/12/28 06:00. doi: 10.1007/978-1-4614-5037-5_14 . - DOI - PubMed
-
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300 . - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

