Splice variants of the extracellular region of RON receptor tyrosine kinase in lung cancer cell lines identified by PCR and sequencing
- PMID: 29121914
- PMCID: PMC5679369
- DOI: 10.1186/s12885-017-3747-x
Splice variants of the extracellular region of RON receptor tyrosine kinase in lung cancer cell lines identified by PCR and sequencing
Abstract
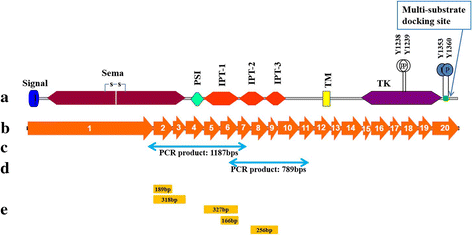
Background: Altered expression of receptor tyrosine kinases (RTKs) is a major driver of growth and metastasis of cancers. Recepteur d'origine nantais (RON) receptor is a single-pass transmembrane RTK aberrantly expressed in a number of cancers. Efforts to block deregulated RON signaling in tumors using small molecule kinase inhibitors or antibodies are complicated by the presence of unknown number/types of isoforms of RON, which, despite having similar sequences, are localized differently and mediate varied functions. The objective of this study was to identify splice variants of RON transcripts between exons 1 and 10 that code for the extracellular region.
Methods: Direct cDNA sequencing was performed for the transcript between exons 1-10 of RON by Sanger sequencing in various lung cancer cell lines.
Results: PCR amplification and bi-directional sequencing of cDNA for section between exons 1 and 10 from lung cancer cell lines revealed the presence of several splice variants of RON transcripts; the variants were formed by skipping of exons 2, 2-3, 5-6, 6 and 8-9. Each of these transcript variants were found in one or more cell lines. While the variants formed by skipping of exons 2, 2-3 and 5-6 resulted in loss of 63, 106 and 109 amino acids, respectively, and didn't cause reading-frameshift, the transcripts formed by skipping of exons 6 and 8-9 caused reading-frameshift. Splice variant lacking exons 8-9 was found in 13 out of 23 cell lines tested.
Conclusion: Lung cancer cell lines contain several splice variants of RON which involve skipping of exons coding for extracellular region. Some of the splicing changes result in reading-frameshift and the N-terminally truncated isoforms are expected to be secreted out. The ubiquitous nature of alternative splicing events in RON suggests the need for isoform specific approaches to functional analysis and therapeutic targeting of RON.
Keywords: Alternative splicing; Lung cancer; Macrophage stimulating protein; RON; RON isoform; Receptor tyrosine kinase.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Identification of the splice variants of Recepteur d'Origine nantais (RON) in lung cancer cell lines.Gene. 2018 Dec 30;679:335-340. doi: 10.1016/j.gene.2018.09.027. Epub 2018 Sep 14. Gene. 2018. PMID: 30223007
-
Novel splicing variants of recepteur d'origine nantais (RON) tyrosine kinase involving exons 15-19 in lung cancer.Lung Cancer. 2016 Feb;92:41-6. doi: 10.1016/j.lungcan.2015.12.002. Epub 2015 Dec 12. Lung Cancer. 2016. PMID: 26775595
-
Recepteur d'Origine nantais (RON) tyrosine kinase splicing variants lacking exons 18 and 19 occur ubiquitously in lung cancer.Int J Clin Exp Med. 2015 Nov 15;8(11):20778-86. eCollection 2015. Int J Clin Exp Med. 2015. PMID: 26885001 Free PMC article.
-
Multiple variants of the RON receptor tyrosine kinase: biochemical properties, tumorigenic activities, and potential drug targets.Cancer Lett. 2007 Nov 18;257(2):157-64. doi: 10.1016/j.canlet.2007.08.007. Epub 2007 Sep 21. Cancer Lett. 2007. PMID: 17889431 Review.
-
Oncogenesis of RON receptor tyrosine kinase: a molecular target for malignant epithelial cancers.Acta Pharmacol Sin. 2006 Jun;27(6):641-50. doi: 10.1111/j.1745-7254.2006.00361.x. Acta Pharmacol Sin. 2006. PMID: 16723080 Review.
Cited by
-
MAGOH promotes gastric cancer progression via hnRNPA1 expression inhibition-mediated RONΔ160/PI3K/AKT signaling pathway activation.J Exp Clin Cancer Res. 2024 Jan 25;43(1):32. doi: 10.1186/s13046-024-02946-8. J Exp Clin Cancer Res. 2024. PMID: 38268030 Free PMC article.
-
An Introduction and Overview of RON Receptor Tyrosine Kinase Signaling.Genes (Basel). 2023 Feb 17;14(2):517. doi: 10.3390/genes14020517. Genes (Basel). 2023. PMID: 36833444 Free PMC article. Review.
-
Alternative Splicing Events and Their Clinical Significance in Colorectal Cancer: Targeted Therapeutic Opportunities.Cancers (Basel). 2023 Aug 7;15(15):3999. doi: 10.3390/cancers15153999. Cancers (Basel). 2023. PMID: 37568815 Free PMC article. Review.
-
Splicing to orchestrate cell fate.Mol Ther Nucleic Acids. 2024 Dec 6;36(1):102416. doi: 10.1016/j.omtn.2024.102416. eCollection 2025 Mar 11. Mol Ther Nucleic Acids. 2024. PMID: 39811494 Free PMC article. Review.
-
Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay.Int J Mol Sci. 2020 Dec 10;21(24):9424. doi: 10.3390/ijms21249424. Int J Mol Sci. 2020. PMID: 33321981 Free PMC article. Review.
References
-
- Gaudino G, Avantaggiato V, Follenzi A, Acampora D, Simeone A, Comoglio PM. The proto-oncogene RON is involved in development of epithelial, bone and neuro-endocrine tissues. Oncogene. 1995;11(12):2627–2637. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

