Quantifying engineered nanomaterial toxicity: comparison of common cytotoxicity and gene expression measurements
- PMID: 29121949
- PMCID: PMC5679359
- DOI: 10.1186/s12951-017-0312-3
Quantifying engineered nanomaterial toxicity: comparison of common cytotoxicity and gene expression measurements
Abstract
Background: When evaluating the toxicity of engineered nanomaterials (ENMS) it is important to use multiple bioassays based on different mechanisms of action. In this regard we evaluated the use of gene expression and common cytotoxicity measurements using as test materials, two selected nanoparticles with known differences in toxicity, 5 nm mercaptoundecanoic acid (MUA)-capped InP and CdSe quantum dots (QDs). We tested the effects of these QDs at concentrations ranging from 0.5 to 160 µg/mL on cultured normal human bronchial epithelial (NHBE) cells using four common cytotoxicity assays: the dichlorofluorescein assay for reactive oxygen species (ROS), the lactate dehydrogenase assay for membrane viability (LDH), the mitochondrial dehydrogenase assay for mitochondrial function, and the Comet assay for DNA strand breaks.
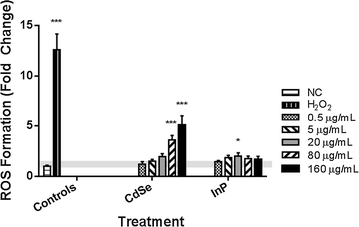
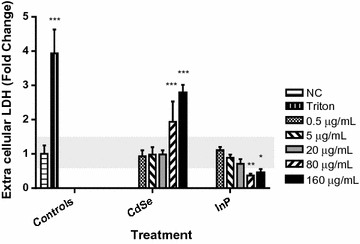
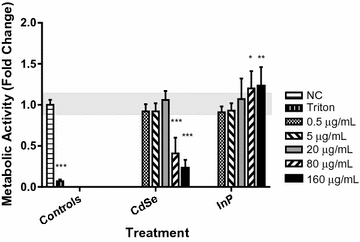
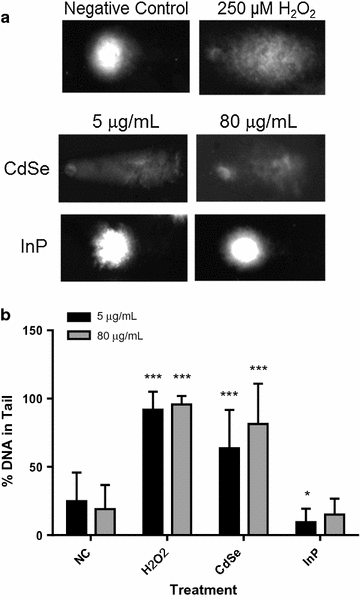
Results: The cytotoxicity assays showed similar trends when exposed to nanoparticles for 24 h at 80 µg/mL with a threefold increase in ROS with exposure to CdSe QDs compared to an insignificant change in ROS levels after exposure to InP QDs, a twofold increase in the LDH necrosis assay in NHBE cells with exposure to CdSe QDs compared to a 50% decrease for InP QDs, a 60% decrease in the mitochondrial function assay upon exposure to CdSe QDs compared to a minimal increase in the case of InP and significant DNA strand breaks after exposure to CdSe QDs compared to no significant DNA strand breaks with InP. High-throughput quantitative real-time polymerase chain reaction (qRT-PCR) data for cells exposed for 6 h at a concentration of 80 µg/mL were consistent with the cytotoxicity assays showing major differences in DNA damage, DNA repair and mitochondrial function gene regulatory responses to the CdSe and InP QDs. The BRCA2, CYP1A1, CYP1B1, CDK1, SFN and VEGFA genes were observed to be upregulated specifically from increased CdSe exposure and suggests their possible utility as biomarkers for toxicity.
Conclusions: This study can serve as a model for comparing traditional cytotoxicity assays and gene expression measurements and to determine candidate biomarkers for assessing the biocompatibility of ENMs.
Keywords: Biomarkers; Comet assay; Cytotoxicity; Gene expression; Genotoxicity; Nanomaterials; Quantum dots.
Figures
References
-
- Borm PJ, Robbins D, Haubold S, Kuhlbusch T, Fissan H, Donaldson K, Schins R, Stone V, Kreyling W, Lademann J, Krutmann J, Warheit D, Oberdorster E. The potential risks of nanomaterials: a review carried out for ECETOC. Part Fibre Toxicol. 2006;3:1–35. doi: 10.1186/1743-8977-3-11. - DOI - PMC - PubMed
-
- Pal BN, Ghosh Y, Brovelli S, Laocharoensuk R, Klimov VI, Hollingsworth JA, Htoon H. Giant CdSe/CdS core/shell nanocrystal quantum dots as efficient electroluminescent materials: strong influence of shell thickness on light-emitting diode performance. Nano Lett. 2012;12:331–336. doi: 10.1021/nl203620f. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

