Genetic Predictors of Response to Systemic Therapy in Esophagogastric Cancer
- PMID: 29122777
- PMCID: PMC5813492
- DOI: 10.1158/2159-8290.CD-17-0787
Genetic Predictors of Response to Systemic Therapy in Esophagogastric Cancer
Abstract
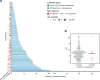
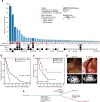
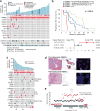
The incidence of esophagogastric cancer is rapidly rising, but only a minority of patients derive durable benefit from current therapies. Chemotherapy as well as anti-HER2 and PD-1 antibodies are standard treatments. To identify predictive biomarkers of drug sensitivity and mechanisms of resistance, we implemented prospective tumor sequencing of patients with metastatic esophagogastric cancer. There was no association between homologous recombination deficiency defects and response to platinum-based chemotherapy. Patients with microsatellite instability-high tumors were intrinsically resistant to chemotherapy but more likely to achieve durable responses to immunotherapy. The single Epstein-Barr virus-positive patient achieved a durable, complete response to immunotherapy. The level of ERBB2 amplification as determined by sequencing was predictive of trastuzumab benefit. Selection for a tumor subclone lacking ERBB2 amplification, deletion of ERBB2 exon 16, and comutations in the receptor tyrosine kinase, RAS, and PI3K pathways were associated with intrinsic and/or acquired trastuzumab resistance. Prospective genomic profiling can identify patients most likely to derive durable benefit to immunotherapy and trastuzumab and guide strategies to overcome drug resistance.Significance: Clinical application of multiplex sequencing can identify biomarkers of treatment response to contemporary systemic therapies in metastatic esophagogastric cancer. This large prospective analysis sheds light on the biological complexity and the dynamic nature of therapeutic resistance in metastatic esophagogastric cancers. Cancer Discov; 8(1); 49-58. ©2017 AACR.See related commentary by Sundar and Tan, p. 14See related article by Pectasides et al., p. 37This article is highlighted in the In This Issue feature, p. 1.
©2017 American Association for Cancer Research.
Figures

Comment in
-
Genomic Analyses and Precision Oncology in Gastroesophageal Cancer: Forwards or Backwards?Cancer Discov. 2018 Jan;8(1):14-16. doi: 10.1158/2159-8290.CD-17-1295. Cancer Discov. 2018. PMID: 29311223
-
The direction of travel to better outcomes for patients with oesophago-gastric cancer.J Thorac Dis. 2018 Mar;10(3):1236-1238. doi: 10.21037/jtd.2018.03.23. J Thorac Dis. 2018. PMID: 29708129 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

