A novel form of RNA double helix based on G·U and C·A+ wobble base pairing
- PMID: 29122970
- PMCID: PMC5769748
- DOI: 10.1261/rna.064048.117
A novel form of RNA double helix based on G·U and C·A+ wobble base pairing
Abstract
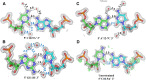
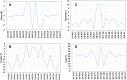
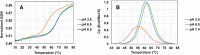
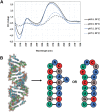
Wobble base pairs are critical in various physiological functions and have been linked to local structural perturbations in double-helical structures of nucleic acids. We report a 1.38-Å resolution crystal structure of an antiparallel octadecamer RNA double helix in overall A conformation, which includes a unique, central stretch of six consecutive wobble base pairs (W helix) with two G·U and four rare C·A+ wobble pairs. Four adenines within the W helix are N1-protonated and wobble-base-paired with the opposing cytosine through two regular hydrogen bonds. Combined with the two G·U pairs, the C·A+ base pairs facilitate formation of a half turn of W-helical RNA flanked by six regular Watson-Crick base pairs in standard A conformation on either side. RNA melting experiments monitored by differential scanning calorimetry, UV and circular dichroism spectroscopy demonstrate that the RNA octadecamer undergoes a pH-induced structural transition which is consistent with the presence of a duplex with C·A+ base pairs at acidic pH. Our crystal structure provides a first glimpse of an RNA double helix based entirely on wobble base pairs with possible applications in RNA or DNA nanotechnology and pH biosensors.
Keywords: RNA double helix; adenine N1 protonation; pH-dependent structural variation; wobble base pairing; wobble helix.
© 2018 Garg and Heinemann; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

Similar articles
-
Crystal structure of r(GUGUGUA)dC with tandem G x U/U x G wobble pairs with strand slippage.J Mol Biol. 1997 Jul 18;270(3):511-9. doi: 10.1006/jmbi.1997.1118. J Mol Biol. 1997. PMID: 9237915
-
Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.RNA Biol. 2020 Mar;17(3):325-334. doi: 10.1080/15476286.2019.1700034. Epub 2019 Dec 18. RNA Biol. 2020. PMID: 31852354 Free PMC article.
-
Structure of an RNA internal loop consisting of tandem C-A+ base pairs.Biochemistry. 1998 Aug 25;37(34):11726-31. doi: 10.1021/bi980758j. Biochemistry. 1998. PMID: 9718295
-
An innate twist between Crick's wobble and Watson-Crick base pairs.RNA. 2013 Aug;19(8):1038-53. doi: 10.1261/rna.036905.112. RNA. 2013. PMID: 23861536 Free PMC article. Review.
-
New information content in RNA base pairing deduced from quantitative analysis of high-resolution structures.Methods. 2009 Mar;47(3):177-86. doi: 10.1016/j.ymeth.2008.12.003. Epub 2009 Jan 14. Methods. 2009. PMID: 19150407 Free PMC article. Review.
Cited by
-
In Silico Study of piRNA Interactions with the SARS-CoV-2 Genome.Int J Mol Sci. 2022 Aug 31;23(17):9919. doi: 10.3390/ijms23179919. Int J Mol Sci. 2022. PMID: 36077317 Free PMC article.
-
miR395e from Manihot esculenta Decreases Expression of PD-L1 in Renal Cancer: A Preliminary Study.Genes (Basel). 2025 Feb 27;16(3):293. doi: 10.3390/genes16030293. Genes (Basel). 2025. PMID: 40149445 Free PMC article.
-
Preclinical development of kinetin as a safe error-prone SARS-CoV-2 antiviral able to attenuate virus-induced inflammation.Nat Commun. 2023 Jan 13;14(1):199. doi: 10.1038/s41467-023-35928-z. Nat Commun. 2023. PMID: 36639383 Free PMC article.
-
Endogenous piRNAs Can Interact with the Omicron Variant of the SARS-CoV-2 Genome.Curr Issues Mol Biol. 2023 Apr 3;45(4):2950-2964. doi: 10.3390/cimb45040193. Curr Issues Mol Biol. 2023. PMID: 37185717 Free PMC article.
-
Unraveling the Enzyme-Substrate Properties for APOBEC3A-Mediated RNA Editing.J Mol Biol. 2023 Sep 1;435(17):168198. doi: 10.1016/j.jmb.2023.168198. Epub 2023 Jul 11. J Mol Biol. 2023. PMID: 37442413 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
