Expression of COBLL1 encoding novel ROR1 binding partner is robust predictor of survival in chronic lymphocytic leukemia
- PMID: 29122990
- PMCID: PMC5792276
- DOI: 10.3324/haematol.2017.178699
Expression of COBLL1 encoding novel ROR1 binding partner is robust predictor of survival in chronic lymphocytic leukemia
Abstract
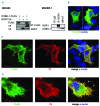
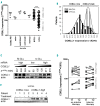
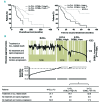
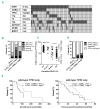
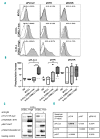
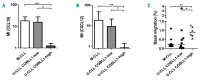
Chronic lymphocytic leukemia is a disease with up-regulated expression of the transmembrane tyrosine-protein kinase ROR1, a member of the Wnt/planar cell polarity pathway. In this study, we identified COBLL1 as a novel interaction partner of ROR1. COBLL1 shows clear bimodal expression with high levels in chronic lymphocytic leukemia patients with mutated IGHV and approximately 30% of chronic lymphocytic leukemia patients with unmutated IGHV. In the remaining 70% of chronic lymphocytic leukemia patients with unmutated IGHV, COBLL1 expression is low. Importantly, chronic lymphocytic leukemia patients with unmutated IGHV and high COBLL1 have an unfavorable disease course with short overall survival and time to second treatment. COBLL1 serves as an independent molecular marker for overall survival in chronic lymphocytic leukemia patients with unmutated IGHV. In addition, chronic lymphocytic leukemia patients with unmutated IGHV and high COBLL1 show impaired motility and chemotaxis towards CCL19 and CXCL12 as well as enhanced B-cell receptor signaling pathway activation demonstrated by increased PLCγ2 and SYK phosphorylation after IgM stimulation. COBLL1 expression also changes during B-cell maturation in non-malignant secondary lymphoid tissue with a higher expression in germinal center B cells than naïve and memory B cells. Our data thus suggest COBLL1 involvement not only in chronic lymphocytic leukemia but also in B-cell development. In summary, we show that expression of COBLL1, encoding novel ROR1-binding partner, defines chronic lymphocytic leukemia subgroups with a distinct response to microenvironmental stimuli, and independently predicts survival of chronic lymphocytic leukemia with unmutated IGHV.
Copyright© 2018 Ferrata Storti Foundation.
Figures
Similar articles
-
COBLL1, LPL and ZAP70 expression defines prognostic subgroups of chronic lymphocytic leukemia patients with high accuracy and correlates with IGHV mutational status.Leuk Lymphoma. 2017 Jan;58(1):70-79. doi: 10.1080/10428194.2016.1180690. Epub 2016 May 17. Leuk Lymphoma. 2017. PMID: 27185377
-
Autocrine Signaling by Wnt-5a Deregulates Chemotaxis of Leukemic Cells and Predicts Clinical Outcome in Chronic Lymphocytic Leukemia.Clin Cancer Res. 2016 Jan 15;22(2):459-69. doi: 10.1158/1078-0432.CCR-15-0154. Epub 2015 Aug 3. Clin Cancer Res. 2016. PMID: 26240275 Free PMC article.
-
Association between immunoglobulin heavy-chain variable region mutational status and isolated favorable baseline genomic aberrations in chronic lymphocytic leukemia.Leuk Lymphoma. 2018 Jan;59(1):59-68. doi: 10.1080/10428194.2017.1323271. Epub 2017 Jun 22. Leuk Lymphoma. 2018. PMID: 28641468 Free PMC article.
-
Why Is the Immunoglobulin Heavy Chain Gene Mutation Status a Prognostic Indicator in Chronic Lymphocytic Leukemia?Acta Haematol. 2018;140(1):51-54. doi: 10.1159/000491382. Epub 2018 Aug 16. Acta Haematol. 2018. PMID: 30114695 Free PMC article. Review.
-
[Impact of immunoglobulin gene somatic high mutation on prognosis of the chronic lymphocytic leukemia].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2009 Dec;17(6):1588-91. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2009. PMID: 20030953 Review. Chinese.
Cited by
-
Multiset multicover methods for discriminative marker selection.Cell Rep Methods. 2022 Nov 11;2(11):100332. doi: 10.1016/j.crmeth.2022.100332. eCollection 2022 Nov 21. Cell Rep Methods. 2022. PMID: 36452867 Free PMC article.
-
Metabolic Effects of the Waist-To-Hip Ratio Associated Locus GRB14/COBLL1 Are Related to GRB14 Expression in Adipose Tissue.Int J Mol Sci. 2022 Aug 2;23(15):8558. doi: 10.3390/ijms23158558. Int J Mol Sci. 2022. PMID: 35955692 Free PMC article.
-
Functional interdependence of the actin nucleator Cobl and Cobl-like in dendritic arbor development.Elife. 2021 Jul 15;10:e67718. doi: 10.7554/eLife.67718. Elife. 2021. PMID: 34264190 Free PMC article.
-
Of Lymph Nodes and CLL Cells: Deciphering the Role of CCR7 in the Pathogenesis of CLL and Understanding Its Potential as Therapeutic Target.Front Immunol. 2021 Mar 24;12:662866. doi: 10.3389/fimmu.2021.662866. eCollection 2021. Front Immunol. 2021. PMID: 33841445 Free PMC article. Review.
-
Radiation-response in primary fibroblasts of long-term survivors of childhood cancer with and without second primary neoplasms: the KiKme study.Mol Med. 2022 Sep 6;28(1):105. doi: 10.1186/s10020-022-00520-6. Mol Med. 2022. PMID: 36068491 Free PMC article.
References
-
- Masiakowski P, Carroll RD. A novel family of cell surface receptors with tyrosine kinase-like domain. J Biol Chem. 1992; 267(36):26181–26190. - PubMed
-
- Baskar S, Kwong KY, Hofer T, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008; 14(2):396–404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

