BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM
- PMID: 29123267
- PMCID: PMC5785741
- DOI: 10.1038/bjc.2017.385
BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM
Abstract
Background: Bactericidal/Permeability-increasing-fold-containing family B member 1 (BPIFB1, previously termed LPLUNC1) is highly expressed in the nasopharynx, significantly downregulated in nasopharyngeal carcinoma (NPC), and associated with prognosis in NPC patients. Because metastasis represents the primary cause of NPC-related death, we explored the role of BPIFB1 in NPC migration and invasion.
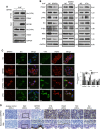
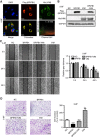
Methods: The role of BPIFB1 in NPC metastasis was investigated in vitro and in vivo. A co-immunoprecipitation assay coupled with mass spectrometry was used to identify BPIFB1-binding proteins. Additionally, western blotting, immunofluorescence, and immunohistochemistry allowed assessment of the molecular mechanisms associated with BPIFB1-specific metastatic inhibition via vitronectin (VTN) and vimentin (VIM) interactions.
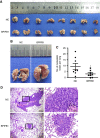
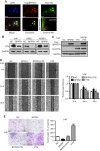
Results: Our results showed that BPIFB1 expression markedly inhibited NPC cell migration, invasion, and lung-metastatic abilities. Additionally, identification of two BPIFB1-interacting proteins, VTN and VIM, showed that BPIFB1 reduced VTN expression and the formation of a VTN-integrin αV complex in NPC cells, leading to inhibition of the FAK/Src/ERK signalling pathway. Moreover, BPIFB1 attenuated NPC cell migration and invasion by inhibiting VTN- or VIM-induced epithelial-mesenchymal transition.
Conclusions: This study represents the first demonstration of BPIFB1 function in NPC migration, invasion, and lung metastasis. Our findings indicate that re-expression of BPIFB1 might represent a useful strategy for preventing and treating NPC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Barnes D, Wolfe R, Serrero G, McClure D, Sato G (1980) Effects of a serum spreading factor on growth and morphology of cells in serum-free medium. J Supramol Struct 14: 47–63. - PubMed
-
- Bingle CD, Craven CJ (2003) Comparative analysis of the PLUNC (palate, lung and nasal epithelium clone) protein families. Biochem Soc Trans 31: 806–809. - PubMed
-
- Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q, Chen P, Shi L, Lian Y, Jing Y, Tang K, Li Z, Zhou Y, Zhou M, Xiang B, Li X, Yang J, Xiong W, Li G, Zeng Z (2015) Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget 6: 20404–20418. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

