Long noncoding RNA and mRNA profiling in MDA-MB-231 cells following RNAi-mediated knockdown of SIRT7
- PMID: 29123410
- PMCID: PMC5661475
- DOI: 10.2147/OTT.S149048
Long noncoding RNA and mRNA profiling in MDA-MB-231 cells following RNAi-mediated knockdown of SIRT7
Abstract
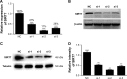
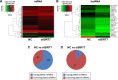
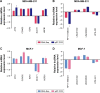
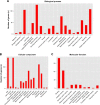
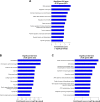
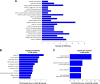
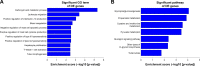
Breast cancer is one of the most common malignant cancers among women and a major clinical obstacle. Although studies have reported the abnormal expression of SIRT7 in breast cancer, whether the function of SIRT7 regulates the expression of long noncoding RNAs (lncRNAs) in breast cancer remains unknown. We aimed to determine the differential expressions of mRNAs and lncRNAs associated with SIRT7 and understand the regulatory mechanism of SIRT7 in breast cancer. RNA sequencing was performed to explore the transcriptome in MDA-MB-231 cells after SIRT7 depletion, and a total of 50,634 different transcripts were identified. In comparison with the negative control, siSIRT7 groups showed 240 differentially expressed mRNAs and 26 differentially expressed lncRNAs. Gene ontology analysis revealed that the differentially expressed mRNAs mainly regulated DNA replication, CXCR chemokine receptor binding, and maturation of large subunit rRNA from tricistronic rRNA transcript, nucleoplasm, mitochondrion, and NAD+ ADP-ribosyltransferase activity. Kyoto Encyclopedia of Genes and Genomes analysis showed that the differentially expressed mRNAs were mainly involved in pathways associated with MAPK signaling pathway, tumor necrosis factor signaling pathway, hepatitis B, and cancer. Moreover, the target genes of the differentially expressed lncRNAs mainly regulated the carboxylic acid metabolic processes and were involved in glycolysis pathway. The mRNA-lncRNA coexpression network comprised 186 mRNAs and 23 lncRNAs. Our results provide essential data regarding differentially expressed lncRNAs and mRNAs after the depletion of SIRT7 in breast cancer cells, which may be useful to elucidate the role of SIRT7 in breast cancer development.
Keywords: RNA-Seq; SIRT7; breast cancer cell; lncRNA; mRNA.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures


Similar articles
-
Differential Expression Profiles of the Transcriptome in Breast Cancer Cell Lines Revealed by Next Generation Sequencing.Cell Physiol Biochem. 2017;44(2):804-816. doi: 10.1159/000485344. Epub 2017 Nov 24. Cell Physiol Biochem. 2017. PMID: 29176322
-
Integrative analysis of the contribution of mRNAs and long non‑coding RNAs to the pathogenesis of asthma.Mol Med Rep. 2019 Sep;20(3):2617-2624. doi: 10.3892/mmr.2019.10511. Epub 2019 Jul 19. Mol Med Rep. 2019. PMID: 31524265 Free PMC article.
-
Comprehensive Analysis of Differentially Expressed Profiles of lncRNAs/mRNAs and miRNAs with Associated ceRNA Networks in Triple-Negative Breast Cancer.Cell Physiol Biochem. 2018;50(2):473-488. doi: 10.1159/000494162. Epub 2018 Oct 11. Cell Physiol Biochem. 2018. PMID: 30308479
-
Systematic Analysis of Long Non-Coding RNAs and mRNAs in the Ovaries of Duroc Pigs During Different Follicular Stages Using RNA Sequencing.Int J Mol Sci. 2018 Jun 11;19(6):1722. doi: 10.3390/ijms19061722. Int J Mol Sci. 2018. PMID: 29891752 Free PMC article.
-
Profile and validation of dysregulated long non‑coding RNAs and mRNAs in ovarian cancer.Oncol Rep. 2018 Nov;40(5):2964-2976. doi: 10.3892/or.2018.6654. Epub 2018 Aug 17. Oncol Rep. 2018. PMID: 30132558
Cited by
-
Underexplored reciprocity between genome-wide methylation status and long non-coding RNA expression reflected in breast cancer research: potential impacts for the disease management in the framework of 3P medicine.EPMA J. 2023 May 22;14(2):249-273. doi: 10.1007/s13167-023-00323-7. eCollection 2023 Jun. EPMA J. 2023. PMID: 37275549 Free PMC article. Review.
-
LINC00160 mediated paclitaxel-And doxorubicin-resistance in breast cancer cells by regulating TFF3 via transcription factor C/EBPβ.J Cell Mol Med. 2020 Aug;24(15):8589-8602. doi: 10.1111/jcmm.15487. Epub 2020 Jul 11. J Cell Mol Med. 2020. PMID: 32652877 Free PMC article.
-
SIRT7 is a Prognostic Biomarker in Kidney Renal Clear Cell Carcinoma That is Correlated with Immune Cell Infiltration.Int J Gen Med. 2022 Mar 19;15:3167-3182. doi: 10.2147/IJGM.S353610. eCollection 2022. Int J Gen Med. 2022. PMID: 35342301 Free PMC article.
-
HNRNPA2B1 is a potential biomarker of breast cancer related to prognosis and immune infiltration.Aging (Albany NY). 2023 Sep 5;15(17):8712-8728. doi: 10.18632/aging.204992. Epub 2023 Sep 5. Aging (Albany NY). 2023. PMID: 37671941 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

