Utilizing Biotinylated Proteins Expressed in Yeast to Visualize DNA-Protein Interactions at the Single-Molecule Level
- PMID: 29123507
- PMCID: PMC5662892
- DOI: 10.3389/fmicb.2017.02062
Utilizing Biotinylated Proteins Expressed in Yeast to Visualize DNA-Protein Interactions at the Single-Molecule Level
Abstract
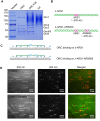
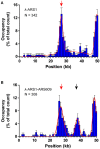
Much of our knowledge in conventional biochemistry has derived from bulk assays. However, many stochastic processes and transient intermediates are hidden when averaged over the ensemble. The powerful technique of single-molecule fluorescence microscopy has made great contributions to the understanding of life processes that are inaccessible when using traditional approaches. In single-molecule studies, quantum dots (Qdots) have several unique advantages over other fluorescent probes, such as high brightness, extremely high photostability, and large Stokes shift, thus allowing long-time observation and improved signal-to-noise ratios. So far, however, there is no convenient way to label proteins purified from budding yeast with Qdots. Based on BirA-Avi and biotin-streptavidin systems, we have established a simple method to acquire a Qdot-labeled protein and visualize its interaction with DNA using total internal reflection fluorescence microscopy. For proof-of-concept, we chose replication protein A (RPA) and origin recognition complex (ORC) as the proteins of interest. Proteins were purified from budding yeast with high biotinylation efficiency and rapidly labeled with streptavidin-coated Qdots. Interactions between proteins and DNA were observed successfully at the single-molecule level.
Keywords: BirA-Avi; DNA-protein interaction; biotin-streptavidin; budding yeast; quantum dots; total internal reflection fluorescence microscopy.
Figures




Similar articles
-
Visualizing the Interaction Between the Qdot-labeled Protein and Site-specifically Modified λ DNA at the Single Molecule Level.J Vis Exp. 2018 Jul 17;(137):57967. doi: 10.3791/57967. J Vis Exp. 2018. PMID: 30080193 Free PMC article.
-
Lipid-Specific Labeling of Enveloped Viruses with Quantum Dots for Single-Virus Tracking.mBio. 2020 May 19;11(3):e00135-20. doi: 10.1128/mBio.00135-20. mBio. 2020. PMID: 32430465 Free PMC article.
-
Limitations of Qdot labelling compared to directly-conjugated probes for single particle tracking of B cell receptor mobility.Sci Rep. 2017 Sep 12;7(1):11379. doi: 10.1038/s41598-017-11563-9. Sci Rep. 2017. PMID: 28900238 Free PMC article.
-
Fluorescent Biosensors Based on Single-Molecule Counting.Acc Chem Res. 2016 Sep 20;49(9):1722-30. doi: 10.1021/acs.accounts.6b00237. Epub 2016 Sep 1. Acc Chem Res. 2016. PMID: 27583695 Review.
-
Meet the neighbors: Mapping local protein interactomes by proximity-dependent labeling with BioID.Proteomics. 2016 Oct;16(19):2503-2518. doi: 10.1002/pmic.201600123. Epub 2016 Jul 27. Proteomics. 2016. PMID: 27329485 Free PMC article. Review.
Cited by
-
Visualizing the Interaction Between the Qdot-labeled Protein and Site-specifically Modified λ DNA at the Single Molecule Level.J Vis Exp. 2018 Jul 17;(137):57967. doi: 10.3791/57967. J Vis Exp. 2018. PMID: 30080193 Free PMC article.
-
Anticipating metastasis through electrochemical immunosensing of tumor hypoxia biomarkers.Anal Bioanal Chem. 2022 Jan;414(1):399-412. doi: 10.1007/s00216-021-03240-8. Epub 2021 Feb 26. Anal Bioanal Chem. 2022. PMID: 33635388
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

