Divergence and Conservative Evolution of XTNX Genes in Land Plants
- PMID: 29123540
- PMCID: PMC5662649
- DOI: 10.3389/fpls.2017.01844
Divergence and Conservative Evolution of XTNX Genes in Land Plants
Abstract
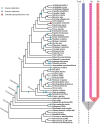
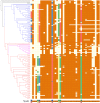
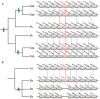
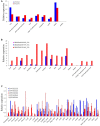
The Toll-interleukin-1 receptor (TIR) and Nucleotide-binding site (NBS) domains are two major components of the TIR-NBS-leucine-rich repeat family plant disease resistance genes. Extensive functional and evolutionary studies have been performed on these genes; however, the characterization of a small group of genes that are composed of atypical TIR and NBS domains, namely XTNX genes, is limited. The present study investigated this specific gene family by conducting genome-wide analyses of 59 green plant genomes. A total of 143 XTNX genes were identified in 51 of the 52 land plant genomes, whereas no XTNX gene was detected in any green algae genomes, which indicated that XTNX genes originated upon emergence of land plants. Phylogenetic analysis revealed that the ancestral XTNX gene underwent two rounds of ancient duplications in land plants, which resulted in the formation of clades I/II and clades IIa/IIb successively. Although clades I and IIb have evolved conservatively in angiosperms, the motif composition difference and sequence divergence at the amino acid level suggest that functional divergence may have occurred since the separation of the two clades. In contrast, several features of the clade IIa genes, including the absence in the majority of dicots, the long branches in the tree, the frequent loss of ancestral motifs, and the loss of expression in all detected tissues of Zea mays, all suggest that the genes in this lineage might have undergone pseudogenization. This study highlights that XTNX genes are a gene family originated anciently in land plants and underwent specific conservative pattern in evolution.
Keywords: XTNX genes; evolution; function divergence; land plants; plant disease resistance genes.
Figures

References
-
- Bailey T. L., Elkan C. (1995). The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3 21–29. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

