Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells
- PMID: 29123954
- PMCID: PMC5665069
- DOI: 10.1080/2162402X.2017.1344804
Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells
Abstract
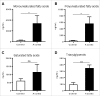
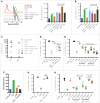
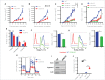
Myeloid-derived suppressor cells (MDSC) promote tumor growth by blocking anti-tumor T cell responses. Recent reports show that MDSC increase fatty acid uptake and fatty acid oxidation (FAO) to support their immunosuppressive functions. Inhibition of FAO promoted a therapeutic T cell-mediated anti-tumor effect. Here, we sought to determine the mechanisms by which tumor-infiltrating MDSC increase the uptake of exogenous lipids and undergo metabolic and functional reprogramming to become highly immunosuppressive cells. The results showed that tumor-derived cytokines (G-CSF and GM-CSF) and the subsequent signaling through STAT3 and STAT5 induce the expression of lipid transport receptors with the resulting increase in the uptake of lipids present at high concentrations in the tumor microenvironment. The intracellular accumulation of lipids increases the oxidative metabolism and activates the immunosuppressive mechanisms. Inhibition of STAT3 or STAT5 signaling or genetic depletion of the fatty acid translocase CD36 inhibits the activation of oxidative metabolism and the induction of immunosuppressive function in tumor-infiltrating MDSC and results in a CD8+ T cell-dependent delay in tumor growth. Of note, human tumor-infiltrating and peripheral blood MDSC also upregulate the expression of lipid transport proteins, and lipids promote the generation of highly suppressive human MDSC in vitro. Our data therefore provide a mechanism by which tumor-derived factors and the high lipid content in the tumor microenvironment can cause the profound metabolic and functional changes found in MDSC and suggest novel approaches to prevent or reverse these processes. These results could further enhance the efficacy of cancer immunotherapy.
Keywords: Immunometabolism; MDSC; immune suppression; lipids; tumor microenvironment.
Figures




References
-
- Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, et al.. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 2016; 7:12150; PMID:27381735; https://doi.org/10.1038/ncomms12150 - DOI - PMC - PubMed
-
- Macatangay BJ, Landay AL, Rinaldo CR. MDSC: A new player in HIV immunopathogenesis. AIDS 2012; 26:1567-9; PMID:22810370; https://doi.org/10.1097/QAD.0b013e328355e682 - DOI - PMC - PubMed
-
- Makarenkova VP, Bansal V, Matta BM, Perez LA, Ochoa JB. CD11b+/Gr-1+ myeloid suppressor cells cause T cell dysfunction after traumatic stress. J Immunol 2006; 176:2085-94; PMID:16455964; https://doi.org/10.4049/jimmunol.176.4.2085 - DOI - PubMed
-
- Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: Expect the unexpected. J Clin Invest 2015; 125:3356-64; PMID:26168215; https://doi.org/10.1172/JCI80005 - DOI - PMC - PubMed
-
- Parker KH, Beury DW, Ostrand-Rosenberg S. Myeloid-derived suppressor cells: Critical cells driving immune suppression in the tumor microenvironment. Adv Cancer Res 2015; 128:95-139; PMID:26216631; https://doi.org/10.1016/bs.acr.2015.04.002 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
