Cold-Shock Domain Family Proteins (Csps) Are Involved in Regulation of Virulence, Cellular Aggregation, and Flagella-Based Motility in Listeria monocytogenes
- PMID: 29124040
- PMCID: PMC5662587
- DOI: 10.3389/fcimb.2017.00453
Cold-Shock Domain Family Proteins (Csps) Are Involved in Regulation of Virulence, Cellular Aggregation, and Flagella-Based Motility in Listeria monocytogenes
Abstract
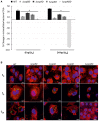
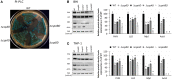
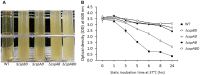
Cold shock-domain family proteins (Csps) are highly conserved nucleic acid binding proteins regulating the expression of various genes including those involved in stress resistance and virulence in bacteria. We show here that Csps are involved in virulence, cell aggregation and flagella-based extracellular motility of Listeria monocytogenes. A L. monocytogenes mutant deleted in all three csp genes (ΔcspABD) is attenuated with respect to human macrophage infection as well as virulence in a zebrafish infection model. Moreover, this mutant is incapable of aggregation and fails to express surface flagella or exhibit swarming motility. An evaluation of double csp gene deletion mutant (ΔcspBD, ΔcspAD and ΔcspAB) strains that produce single csp genes showed that there is redundancy as well as functional differences among the three L. monocytogenes Csps in their contributions to virulence, cellular aggregation, flagella production, and swarming motility. Protein and mRNA expression analysis further showed impaired expression of key virulence and motility genes in the csp mutants. Our observations at protein and mRNA level suggest Csp-dependent expression regulation of these genes at transcriptional and post-transcriptional levels. In a mutant lacking all csp genes (ΔcspABD) as well as those possessing single csp genes (ΔcspBD, ΔcspAD, and ΔcspAB) we detected reduced levels of proteins or activity as well as transcripts from the prfA, hly, mpl, and plcA genes suggesting a Csp-dependent transcriptional regulation of these genes. These csp mutants also had reduced or completely lacked ActA proteins and cell surface flagella but contained elevated actA and flaA mRNA levels compared to the parental wild type strain suggesting Csp involvement in post-transcriptional regulation of these genes. Overall, our results suggest that Csps contribute to the expression regulation of virulence and flagella-associated genes thereby promoting host pathogenicity, cell aggregation and flagella-based motility processes in L. monocytogenes.
Keywords: Csps; Listeria monocytogenes; cellular aggregation; flagella; swarming motility; virulence.
Figures

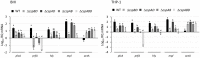

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

